Morphological Profiling vs. Immunocytochemistry: A Modern Paradigm for Neural Culture Validation in Research and Therapy
This article provides a comprehensive comparison of morphological profiling and immunocytochemistry for validating neural cultures, a critical step for ensuring reproducibility in neuroscience research and safety in cell-based therapies.
Morphological Profiling vs. Immunocytochemistry: A Modern Paradigm for Neural Culture Validation in Research and Therapy
Abstract
This article provides a comprehensive comparison of morphological profiling and immunocytochemistry for validating neural cultures, a critical step for ensuring reproducibility in neuroscience research and safety in cell-based therapies. It explores the foundational principles of both techniques, detailing advanced methodological applications from high-content live-cell imaging to AI-based analysis. The content addresses common troubleshooting scenarios and optimization strategies, culminating in a direct validation and comparative analysis of the techniques' accuracy, throughput, and cost-effectiveness. Aimed at researchers, scientists, and drug development professionals, this review synthesizes current evidence to guide the selection and integration of these quality control methods for specific applications in basic research, drug screening, and clinical therapy development.
The Core Principles: Understanding Morphological Profiling and Immunocytochemistry for Neural Cells
Defining Neural Culture Validation and Its Impact on Experimental Reproducibility
The use of human neural cultures, particularly those derived from induced pluripotent stem cells (iPSCs), has revolutionized neuroscience research and drug development by providing physiologically relevant human models. However, genetic drift, clonal heterogeneity, and variations in differentiation protocols cause significant variability in the resulting cell cultures [1]. This heterogeneity directly impacts experimental reproducibility, as the composition, purity, and maturity of neural cultures profoundly affect gene expression and functional activity [1]. Neural culture validation has therefore emerged as an essential practice to ensure reliability in both basic research and clinical applications.
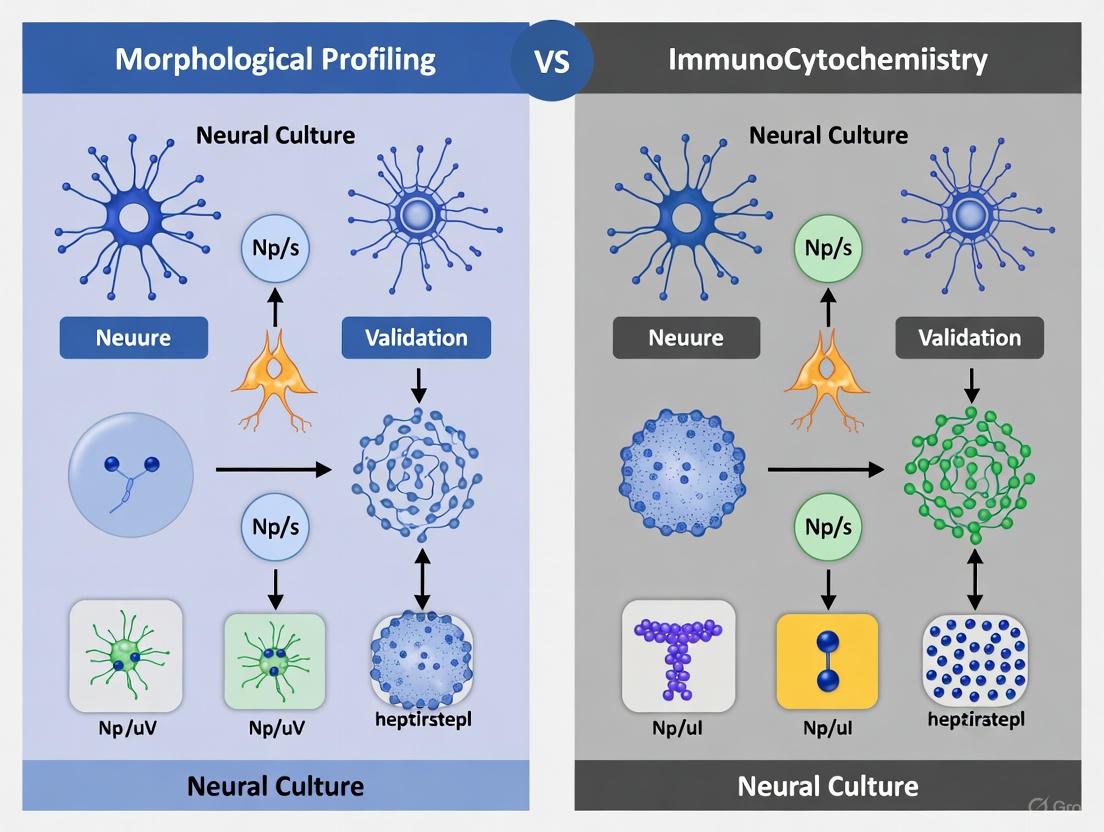
Two principal methodologies have developed for validating neural cultures: traditional immunocytochemistry and emerging morphological profiling approaches. Immunocytochemistry relies on specific antibody-based detection of molecular markers, while morphological profiling uses high-content imaging and computational analysis to identify cell types based on physical characteristics. This guide provides an objective comparison of these methodologies, examining their performance characteristics, experimental requirements, and impacts on experimental reproducibility.
Methodological Comparison: Immunocytochemistry vs. Morphological Profiling
Fundamental Principles and Technical Basis
Immunocytochemistry (ICC) is a well-established technique that utilizes antibody-antigen interactions to detect specific molecular markers. In neural culture validation, researchers use antibodies targeting cell-type-specific proteins to identify and quantify different neural cell types. For example, studies commonly target markers such as PGP 9.5 for general neuronal identification, ChAT for cholinergic neurons, and nNOS for nitrergic neurons [2]. The technique provides direct molecular evidence of cell identity through fluorescent or colorimetric detection.
Morphological Profiling represents a paradigm shift in validation methodology. This approach uses high-content imaging with simple organic dyes combined with computational analysis, including convolutional neural networks (CNNs), to recognize cell types based on their morphotextural fingerprints [1]. Unlike ICC, morphological profiling does not require prior knowledge of specific molecular markers and can distinguish cell types based solely on physical characteristics such as shape, size, texture, and spatial patterns.
Performance Metrics and Validation Outcomes
Table 1: Quantitative Performance Comparison of Validation Methods
| Performance Metric | Immunocytochemistry | Morphological Profiling |
|---|---|---|
| Classification Accuracy | Not systematically reported | 96% (astroglioma/neuroblastoma) [1] |
| Multiplexing Capacity | Limited by antibody host species and spectral overlap | Essentially unlimited through multichannel imaging |
| Throughput | Low to moderate (manual scoring) | High (automated image analysis) |
| Cost per Sample | $50-200 (antibody-dependent) | $10-50 (dyes and computational) |
| Temporal Resolution | Endpoint measurements only | Potential for live-cell monitoring |
| Susceptibility to Density Effects | Minimal | Minimal with regional restriction approach [1] |
Table 2: Application-Specific Performance Characteristics
| Validation Context | Immunocytochemistry Performance | Morphological Profiling Performance |
|---|---|---|
| Basic Cell Type Identification | High specificity and reliability [2] | 96% accuracy in controlled conditions [1] |
| Dense Mixed Cultures | Challenging due to antibody penetration issues | Maintains >96% accuracy with nuclear-focused approach [1] |
| State Determination | Excellent for predefined activation markers | Lower accuracy (78%) for microglial activation states [1] |
| iPSC-Differentiation QC | Standard approach but destructive | 96% vs. 86% for population-level classification [1] |
Impact on Experimental Reproducibility
The reproducibility crisis in neuroscience extends to neural culture studies, where variability in culture composition directly affects experimental outcomes. Immunocytochemistry has served as the gold standard but faces limitations in standardization due to batch-to-batch antibody variations, fixation differences, and subjective scoring [1]. Morphological profiling offers advantages in standardization through quantitative, algorithm-driven classification that minimizes human bias.
Recent initiatives addressing reproducibility challenges, such as the NERVE-ML checklist for machine learning in neural engineering, emphasize the importance of proper validation procedures [3]. The checklist provides guidelines to ensure that computational approaches, including those used in morphological profiling, lead to valid scientific conclusions through appropriate validation strategies [3]. Large-scale collaborative efforts like the International Brain Laboratory's brain-wide mapping study demonstrate the critical importance of standardized validation methods across laboratories [4].
Experimental Protocols for Method Implementation
Immunocytochemistry Protocol for Neural Culture Validation
Sample Preparation:
- Culture neural cells on sterile glass coverslips at appropriate density
- Fix cells with Formal FIXX or 4% paraformaldehyde for 30 minutes
- Permeabilize with 0.05% Triton X-100 in PBS for 30 minutes
- Block with 1% BSA in PBS for 1 hour to reduce nonspecific binding
Antibody Staining:
- Incubate with primary antibodies diluted in blocking buffer (2 hours at room temperature or overnight at 4°C)
- Common primary antibodies: PGP 9.5 (1:1000), Milli-Mark FluoroPan (1:100), ChAT (1:100), nNOS (1:1000) [2]
- Wash 3×5 minutes with PBS/0.05% Triton X-100/1% BSA
- Incubate with species-appropriate secondary antibodies conjugated to fluorophores (1:200, 30 minutes at room temperature)
- Counterstain nuclei with DAPI (1:5000, 5 minutes)
- Mount with Fluoromount and seal with nail polish
Imaging and Analysis:
- Image using fluorescent or confocal microscopy (e.g., Zeiss LSM-710)
- Quantify positive cells manually or using automated counting algorithms
- Express results as percentage of total cells (DAPI-positive) for each marker
Morphological Profiling Protocol for Neural Culture Validation
Cell Staining and Imaging:
- Culture cells in appropriate vessel for high-content imaging
- Stain with cell painting cocktail: 4-channel confocal imaging with dyes targeting nucleus, nucleoli, cytoplasm, Golgi, and actin [1]
- Fix cells if endpoint analysis required, or use live-cell compatible dyes
- Image using high-content microscope with consistent settings across samples
Image Analysis and Classification:
- Segment individual cells using convolutional neural networks
- Extract morphotextural features describing shape, intensity, and texture
- For dense cultures, use nuclear region of interest with immediate periphery to maintain accuracy [1]
- Train classification algorithm (e.g., ResNet CNN) on reference datasets
- Validate classifier performance using separate test dataset
- Apply trained model to new cultures for cell type identification
Validation and Quality Control:
- Compare classification results with immunocytochemistry for benchmark cultures
- Establish confidence thresholds for classification accuracy
- Implement tiered strategy for distinguishing challenging cell states [1]
Visualization of Method Workflows
Immunocytochemistry Workflow
Immunocytochemistry Workflow: Sequential process from cell preparation to analysis.
Morphological Profiling Workflow
Morphological Profiling Workflow: Integrated experimental and computational steps.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential Research Reagents for Neural Culture Validation
| Reagent/Material | Function | Application in ICC | Application in Morph Profiling |
|---|---|---|---|
| Primary Antibodies (PGP 9.5, ChAT, nNOS) | Specific detection of neuronal markers | Essential for target identification | Not required |
| Fluorophore-conjugated Secondary Antibodies | Signal amplification and detection | Required for visualization | Not required |
| Cell Painting Dye Cocktail | Multiplexed staining of cellular compartments | Not used | Essential for morphological feature extraction |
| Fixation Reagents (Paraformaldehyde) | Cellular structure preservation | Required for most protocols | Optional (live-cell compatible) |
| Permeabilization Agents (Triton X-100) | Membrane permeabilization for antibody access | Required for intracellular targets | Not required |
| Blocking Serum (BSA) | Reduction of nonspecific antibody binding | Essential for signal-to-noise optimization | Not required |
| High-Content Imaging System | Automated multiparameter image acquisition | Optional | Essential for data generation |
| Computational Analysis Software | Image analysis and classification | Basic for quantification | Essential for classification |
The validation of neural cultures represents a critical step in ensuring experimental reproducibility across neuroscience research and drug development. While immunocytochemistry provides highly specific, target-driven validation with established protocols, morphological profiling offers a complementary approach with advantages in scalability, standardization, and potentially lower cost. The choice between methodologies depends on specific research requirements, including the need for specific molecular information, throughput requirements, and available expertise.
Future developments in neural culture validation will likely focus on increasing integration between these approaches, leveraging the molecular specificity of immunocytochemistry with the computational power of morphological profiling. Advanced human brain models such as miBrains, which integrate all major brain cell types, will require increasingly sophisticated validation approaches [5]. Similarly, machine learning approaches for classifying neuronal subtypes in co-culture systems demonstrate the potential for computational methods to enhance traditional validation paradigms [6]. As the field progresses, standardized validation frameworks will be essential for improving reproducibility and accelerating the development of novel therapeutics for neurological disorders.
Immunocytochemistry (ICC) is a foundational laboratory technique that employs antibodies to detect and visualize specific antigens, such as proteins, within individual cells, enabling precise localization at a subcellular level through microscopy [7] [8]. This technique is particularly adapted for isolated cells, cell cultures, smears, or fluid-based specimens, distinguishing it from immunohistochemistry (IHC), which is used for tissue sections [9] [8]. In the context of neural culture validation research, ICC has been an indispensable tool for characterizing cell types, confirming neuronal differentiation, and validating the expression of specific biomarkers, such as those found on neural progenitor cells [10] [11]. As the field advances, traditional ICC is increasingly compared with novel, label-free methods like morphological profiling for quality control in complex cultures, framing a modern debate on validation strategies for neuroscience research [1].
Historical Context and Development
The genesis of immunocytochemistry traces back to the pioneering work of Albert H. Coons and colleagues in the early 1940s, who developed the first fluorescent antibody technique to detect pneumococcal antigens in mouse lung tissue [8] [12]. This breakthrough, born from Coons's work on rheumatic fever, marked the birth of immunofluorescence and established the core principle of using antibodies as specific detection tools [12].
Key milestones in the evolution of ICC include:
- 1950s: Adaptation of the technique for isolated cells and cultured cell preparations, shifting from tissue sections to cytological analysis [8].
- 1960s-1970s: Introduction of enzyme-linked antibodies to overcome limitations of fluorescence, such as photobleaching, enabling stable, light-microscopy-compatible signals [8]. A pivotal advancement was the 1970 development of the peroxidase-antiperoxidase (PAP) method by Ludwig A. Sternberger, which significantly amplified signal sensitivity [8].
- 1975: A revolution in specificity occurred with the development of monoclonal antibody production using hybridoma technology by Georges Köhler, César Milstein, and Niels K. Jerne [12]. This provided researchers with a consistent supply of highly specific antibodies targeting single epitopes, drastically improving reproducibility and reducing background noise [8] [12].
- Late 20th Century: Integration of ICC with confocal and super-resolution microscopy significantly enhanced resolution and 3D imaging capabilities, allowing for detailed study of subcellular structures [12].
Core Principles of Immunocytochemistry
The fundamental principle of ICC is the highly specific interaction between an antibody and its target antigen, a concept rooted in Paul Ehrlich's "side-chain theory" [12]. The target antigen, which can be a protein, carbohydrate, or lipid, presents a small region called an epitope that is recognized by the antigen-binding fragment (Fab) region of the antibody [8]. The strength of this interaction is governed by affinity—the intrinsic binding energy of a single Fab-epitope interaction—and avidity, which describes the enhanced overall binding strength from multivalent interactions, such as a bivalent IgG antibody binding multiple epitopes [8].
To visualize this binding, antibodies are conjugated to detectable markers. The two primary detection methods are:
- Immunofluorescence (IF): Uses fluorophore-conjugated antibodies. Signals are detected using fluorescence microscopy [9] [12]. This method allows for multiplexing and high-resolution imaging.
- Chromogenic Detection: Uses enzyme-conjugated antibodies (e.g., horseradish peroxidase) that catalyze a reaction producing a colored precipitate, visible under a light microscope [9] [8]. This method produces a permanent stain.
Table 1: Key Components of an Immunocytochemistry Experiment
| Component | Function | Common Examples |
|---|---|---|
| Primary Antibody | Binds specifically to the antigen of interest. | Monoclonal or polyclonal antibodies from rabbit, mouse. |
| Secondary Antibody | Binds to the primary antibody; conjugated to a marker for detection. | Anti-rabbit IgG conjugated to a fluorophore (e.g., Alexa Fluor 488) or an enzyme (e.g., HRP). |
| Fluorophore | A fluorescent dye that emits light upon excitation. | FITC, TRITC, Cy5, Alexa Fluor dyes [9] [12]. |
| Fixative | Preserves cellular structure and immobilizes antigens. | Paraformaldehyde [9] [13]. |
| Permeabilization Agent | Disrupts the plasma membrane to allow antibody access to intracellular antigens. | Detergents like Triton X-100 [12]. |
| Blocking Buffer | Reduces nonspecific antibody binding to off-target sites. | Serum (e.g., normal goat serum) or protein solutions (e.g., BSA) [9]. |
Direct vs. Indirect Detection Methods
Two main antibody methods are used in ICC, each with distinct advantages and disadvantages [9].
- Direct Method: The primary antibody is directly conjugated to a detectable marker. This method is faster, involves fewer steps, and avoids potential cross-reactivity from secondary antibodies. However, it generally has lower sensitivity and fewer commercially available conjugated primary antibodies [9].
- Indirect Method: An unlabeled primary antibody binds to the antigen, and a labeled secondary antibody that recognizes the primary antibody is then applied. This method offers higher sensitivity because multiple secondary antibodies can bind to a single primary antibody, thereby amplifying the signal. It also provides great flexibility, as one type of labeled secondary antibody can be used with various primary antibodies from the same species [9].
Table 2: Comparison of Direct and Indirect Immunocytochemistry Methods
| Parameter | Direct ICC | Indirect ICC |
|---|---|---|
| Primary Antibody | Conjugated with a detectable marker | Unconjugated |
| Secondary Antibody | Not required | Required; conjugated with a marker |
| Processing Time | Fast (one-step incubation) | Slow (two-step incubation) |
| Sensitivity | Low | High (due to signal amplification) |
| Signal Amplification | No | Yes |
| Commercial Availability | Limited | Many options available |
| Cross-reactivity | Avoided | Must use primary and secondary antibodies from different species |
Diagram 1: Direct vs. Indirect ICC Workflows. The direct method uses a single conjugated antibody, while the indirect method uses a secondary antibody for signal amplification.
Experimental Protocols and Methodologies
A standard ICC protocol involves a series of critical steps designed to preserve cellular architecture, ensure specific antibody binding, and generate a strong, detectable signal while minimizing background [12].
Detailed ICC Protocol for Neural Cultures
The following protocol is synthesized from methodologies used in recent neural culture studies [10] [13] [11]:
- Cell Seeding and Culture: Plate cells (e.g., primary cortical neurons, iPSC-derived neural progenitors) on sterile, poly-l-ornithine-coated glass coverslips placed in a multi-well culture dish. Culture cells in appropriate medium (e.g., MEM Alpha GlutaMAX supplemented with B27 and serum) until they reach the desired confluency, typically not exceeding 50-70% to prevent differentiation [10] [13].
- Fixation: Aspirate the culture medium and wash cells gently with phosphate-buffered saline (PBS). Fix cells by incubating in a 4% paraformaldehyde (PFA) solution in PBS for 10-15 minutes at room temperature [13]. This step cross-links and preserves the cellular structures.
- Permeabilization and Blocking: Remove PFA and wash cells with PBS. Permeabilize the fixed cells by incubating with a detergent solution (e.g., 0.1-0.3% Triton X-100 in PBS) for 5-15 minutes. Subsequently, incubate cells in a blocking buffer (e.g., 5% normal goat serum in PBS) for at least 30 minutes to block nonspecific binding sites [13] [12].
- Antibody Incubation:
- Primary Antibody: Dilute the specific primary antibody (e.g., anti-CREB, anti-PAX6, anti-NESTIN) in an appropriate dilution buffer (often the same as the blocking buffer). Apply the solution to the coverslip and incubate in a humidified chamber. Incubation conditions can vary from 1-2 hours at room temperature to overnight at 4°C [13].
- Washing: After incubation, wash the coverslip thoroughly several times with PBS (e.g., 3 x 5 minutes) to remove unbound primary antibodies.
- Secondary Antibody: Apply a fluorophore-conjugated secondary antibody (e.g., Alexa Fluor 488-conjugated goat anti-rabbit IgG), diluted in buffer, and incubate for 1 hour at room temperature in the dark to protect the fluorophore from light [13].
- Counterstaining and Mounting: Wash the coverslip again with PBS to remove unbound secondary antibody. Incubate with a nuclear counterstain such as DAPI (4',6-diamidino-2-phenylindole) for a few minutes, followed by a final PBS wash [9] [12]. Mount the coverslip onto a glass microscope slide using an antifade mounting medium (e.g., VECTASHIELD) to reduce photobleaching [9].
- Imaging and Analysis: Visualize the stained cells using a fluorescence or confocal microscope. Acquire images and analyze fluorescence intensity and localization using quantitative image analysis software [13] [1].
Exemplar Experimental Application: Validating CRISPR/Cas9 in Neurons
A 2017 study effectively demonstrated the utility of ICC in validating gene disruption in primary neurons [13]. The researchers aimed to disrupt the Creb gene in mouse cortical neurons using the CRISPR/Cas9 system.
- Methodology: They transfected cortical neurons with a CRISPR/Cas9 plasmid targeting Creb. After allowing time for gene editing and protein turnover, the cells were fixed and processed for ICC using an antibody against CREB. Fluorescence intensity was quantified to measure CREB expression.
- Results and Data: Quantitative ICC analysis revealed that CREB expression was abolished in a subset of the transfected neurons, confirming successful gene disruption at a single-cell level. This was further correlated with a decrease in FOS, a downstream target of CREB, and a reduction in dendritic arborization, linking the genetic manipulation to a functional phenotype [13].
- Significance: This protocol showcases how ICC, combined with fluorescence imaging, provides an efficient and reliable method to identify and study mutant neurons in a heterogeneous primary culture, bypassing the need for laborious single-cell genomic sequencing.
Performance Comparison: Antibodies and Reagents
The reliability of ICC data is critically dependent on the performance of antibodies. Independent, open-science characterization efforts like those by YCharOS provide valuable comparative data.
Table 3: Antibody Performance Across Common Applications (Based on YCharOS Data) [14]
| Vendor / Antibody Type | Western Blot Pass Rate | Immunoprecipitation Pass Rate | ICC Pass Rate |
|---|---|---|---|
| Abcam (Recombinant Monoclonal) | 97% | 55% | 83% |
| Abcam (All Antibodies, Average) | 87% | 51% | 66% |
| Industry Competitors (Average) | Lower than Abcam | Lower than Abcam | Lower than Abcam |
The data underscores that recombinant antibodies, which are sequence-defined and produced with high consistency between batches, generally show superior performance, with a pass rate in ICC that can be up to 30% higher than that of polyclonal antibodies [14]. This highlights the importance of antibody validation for specific applications to ensure experimental reproducibility.
ICC vs. Morphological Profiling for Neural Culture Validation
The central nervous system's complexity demands robust validation for in vitro models. While ICC has been the gold standard, emerging label-free methods like morphological profiling present a complementary approach.
A 2024 study directly addressed this by using high-content imaging and convolutional neural networks (CNNs) to identify cell types in dense, mixed neural cultures based solely on their morphological "fingerprint" [1]. This Cell Painting (CP) approach achieved a classification accuracy above 96% for distinguishing different neural cell lines and for evaluating the differentiation status of iPSC-derived neural cultures [1].
Table 4: Immunocytochemistry vs. Morphological Profiling for Neural Culture Validation
| Aspect | Immunocytochemistry (ICC) | Morphological Profiling / Cell Painting |
|---|---|---|
| Basis of Detection | Specific antibody-antigen binding [8]. | Label-free analysis of cellular morphology and texture [1]. |
| Throughput | Medium (requires staining and washing steps). | High (automated, uses simple organic dyes) [1]. |
| Cost per Sample | Higher (cost of antibodies). | Lower (cost of dyes) [1]. |
| Multiplexing Capacity | Excellent for 2-4 targets with spectral separation [9] [12]. | Simultaneously captures thousands of morphological features [1]. |
| Specificity | High (targets specific proteins). | Lower (identifies cell types based on phenotype, not specific markers). |
| Primary Application | Target validation, subcellular localization, pathway analysis. | Quality control, phenotyping, mode-of-action studies [1]. |
| Cell State Sensitivity | Detects molecular expression levels. | Can distinguish activated vs. non-activated microglia states [1]. |
| Key Advantage | High molecular specificity and well-established protocols. | Fast, affordable, scalable, and non-destructive [1]. |
Diagram 2: Comparative Workflows: ICC vs. Morphological Profiling. The two methods represent different approaches, with ICC being targeted and destructive, and morphological profiling being holistic and potentially more scalable.
The study concluded that while ICC provides definitive molecular identification, morphological profiling offers a powerful, complementary means to quantify cell composition in complex mixed neural cultures quickly and cost-effectively, holding great promise for the quality control of iPSC-derived models [1].
The Scientist's Toolkit: Essential Research Reagents
Successful ICC experiments rely on a suite of carefully selected reagents. The following table details key solutions and their functions in a typical ICC workflow.
Table 5: Essential Research Reagent Solutions for ICC
| Reagent / Solution | Function in the Protocol | Key Considerations |
|---|---|---|
| Paraformaldehyde (PFA) 4% | Cross-links proteins to preserve cellular structure and immobilize antigens during fixation [13]. | Concentration and fixation time must be optimized to balance antigen preservation and epitope masking. |
| Triton X-100 (0.1-0.3%) | A detergent that permeabilizes the fixed cell membrane, allowing antibodies to access intracellular targets [12]. | Over-permeabilization can damage cellular structures. |
| Normal Serum (e.g., Goat, Donkey) 5-10% | Used in blocking buffer to occupy nonspecific binding sites, thereby reducing background staining [9] [13]. | Should be from the same species as the host of the secondary antibody for optimal blocking. |
| Primary Antibody Diluent | Buffer (often PBS with serum or BSA) used to dilute the primary antibody to its working concentration. | Stabilizes the antibody and can include preservatives for long-term storage. |
| Fluorophore-Conjugated Secondary Antibody | Binds to the primary antibody and provides the detectable signal for visualization under a microscope [9]. | Must be raised against the host species of the primary antibody and chosen to avoid spectral overlap in multiplexing. |
| Antifade Mounting Medium | Preserves the fluorescence signal by reducing photobleaching during storage and imaging [9]. | Products like VECTASHIELD are commonly used. May include DAPI for nuclear counterstaining. |
Immunocytochemistry has cemented its role as a cornerstone technique in cellular neuroscience, providing unparalleled specificity for protein localization and validation within neural cells. From its origins in the work of Coons to the modern application of highly validated recombinant antibodies, the principles of antibody-antigen binding have remained constant, while the tools and applications have dramatically advanced. In the evolving landscape of neural culture validation, ICC does not stand alone. It is increasingly complemented by high-throughput, label-free methods like morphological profiling, which leverage artificial intelligence to classify cells based on phenotype. The future of neural culture validation lies not in choosing one method over the other, but in strategically integrating the molecular specificity of ICC with the scalable, holistic profiling of morphological analysis to create robust, reproducible, and physiologically relevant models for neuroscience research and drug discovery.
The quest for physiologically relevant human neural models, driven by induced pluripotent stem cell (iPSC) technology, has revolutionized neuroscience research and preclinical drug screening [1]. However, this advancement brings a critical challenge: the need for robust, scalable methods to characterize the composition, purity, and maturity of the resulting complex mixed neural cultures. Traditional validation methods like immunocytochemistry (ICC), while highly specific, are often low in throughput, costly, and destructive, hindering their use in systematic screening pipelines [1]. In response, morphological profiling has emerged as a powerful, unbiased alternative. By combining high-content imaging with artificial intelligence (AI), this approach quantitatively analyzes cellular structure and organization, offering a fast, affordable, and information-rich method for quality control. This guide objectively compares the performance of modern AI-powered morphological profiling against traditional immunocytochemistry for validating neural cultures, providing researchers with the data needed to select the optimal method for their application.
Technical Comparison: Morphological Profiling vs. Immunocytochemistry
The following table summarizes the core performance characteristics of each method.
| Feature | AI-Powered Morphological Profiling | Traditional Immunocytochemistry (ICC) |
|---|---|---|
| Core Principle | AI-driven analysis of cellular morphology from multiplexed dye staining [1] [15] | Targeted visualization of specific antigens using antibody binding |
| Multiplexing Capacity | High (5-6 channels capturing multiple organelles) [1] [15] | Moderate, limited by antibody host species and fluorophore spectra |
| Throughput | High (automated, suitable for large-scale screening) [1] [16] | Low to moderate (often manual and time-consuming) [1] |
| Quantitative Output | High-dimensional (1,000s of features per cell) [1] [15] | Primarily semi-quantitative (e.g., intensity, cell counts) |
| Key Metric: Classification Accuracy | 96% (cell type identification in mixed neural cultures) [1] | Highly variable; depends on antibody specificity and validation |
| Key Metric: Predictive Accuracy | 88-92% (predicting clinical drug response) [16] | Not typically used for predictive functional assessment |
| Cost per Sample | Lower after initial setup | Recurring costs for antibodies |
| Assay Destructiveness | Non-destructive (compatible with live-cell dyes) | Destructive (requires cell fixation) |
Experimental Protocols and Workflows
Protocol for AI-Powered Morphological Profiling (Cell Painting/NeuroPainting)
The Cell Painting assay and its neural-optimized derivative, NeuroPainting, form the basis for most modern morphological profiling workflows [1] [15].
- Step 1: Cell Staining. Cells are stained with a panel of fluorescent dyes to label key cellular compartments:
- Step 2: High-Content Imaging. Stained plates are imaged using automated confocal or high-content microscopes (e.g., Perkin Elmer Phenix) with a 20x or 40x objective [1] [15].
- Step 3: Image Analysis and Feature Extraction. Images are processed using pipelines in software like CellProfiler [15] or with deep learning models (Convolutional Neural Networks) [1]. For each cell, thousands of morphotextural features are extracted, describing the shape, size, intensity, and texture of each labeled compartment [1] [15].
- Step 4: Data Analysis and Classification. The high-dimensional data is analyzed using machine learning. Random Forest classifiers or CNNs are trained to identify cell types or states based on their morphological fingerprints [1] [16].
Protocol for Immunocytochemistry (ICC)
- Step 1: Cell Fixation and Permeabilization. Cultures are fixed with paraformaldehyde (e.g., 4% for 20 minutes) and permeabilized with a detergent like Triton X-100 to allow antibody entry [17].
- Step 2: Blocking. Cells are incubated with a protein block (e.g., 2% fetal bovine serum) to prevent non-specific antibody binding [17].
- Step 3: Antibody Incubation. Cells are incubated with primary antibodies against specific neuronal markers (e.g., MAP2 for mature neurons, GFAP for astrocytes, Synapsin for synapses) [17], followed by fluorescently conjugated secondary antibodies.
- Step 4: Imaging and Analysis. Cells are imaged by epifluorescence or confocal microscopy. Analysis involves manual counting or semi-automated measurement of fluorescence intensity to determine the proportion of cells expressing the target markers.
The Scientist's Toolkit: Essential Research Reagents and Materials
The table below details key reagents and instruments essential for implementing morphological profiling.
| Item Name | Function/Description | Example Use Case |
|---|---|---|
| Hoechst 33342 | Fluorescent stain that binds to DNA in the nucleus. | Segmentation of individual cells and nuclear morphology analysis [15]. |
| Phalloidin (Conjugated) | Binds to filamentous actin (F-actin) in the cytoplasm. | Visualizing and quantifying cytoskeletal organization and cell shape [16] [15]. |
| MitoTracker Dyes | Cell-permeant dyes that accumulate in active mitochondria. | Assessment of mitochondrial mass, distribution, and network health [15]. |
| Wheat Germ Agglutinin (WGA) | Labels glycoproteins and glycolipids on the plasma membrane and Golgi. | Delineating cell boundaries and profiling Golgi apparatus structure [1]. |
| Concanavalin A | Binds to glycoproteins in the endoplasmic reticulum and Golgi. | Characterizing the organization of the secretory pathway [1]. |
| Automated Confocal Microscope | High-content imaging system for automated multi-channel acquisition. | Generating high-quality, high-throughput image data from multi-well plates [15]. |
| CellProfiler Software | Open-source platform for creating customized image analysis pipelines. | Extracting thousands of morphological features from segmented images [15]. |
| Convolutional Neural Network (CNN) | Class of deep learning algorithm, particularly effective for image analysis. | Classifying cell types with high accuracy based on raw image data [1]. |
Performance Analysis: Supporting Experimental Data
Accuracy and Predictive Power in Validation
Independent studies demonstrate the high accuracy of morphological profiling in neural applications. One study using a Cell Painting assay with a CNN achieved above 96% accuracy in classifying neuroblastoma and astrocytoma cell lines in mixed cultures, significantly outperforming a classification based solely on time in culture (86%) [1]. Critically, this high fidelity was maintained even in dense, confluent cultures by focusing on the nuclear region and its immediate environment [1].
Beyond simple identification, morphological profiling shows remarkable predictive power for clinical outcomes. A study on T cells from multiple sclerosis patients used a high-content imaging pipeline and a Random Forest model to predict patient response to natalizumab therapy with 92% accuracy in a discovery cohort and 88% in a validation cohort [16]. This demonstrates that subtle, pretreatment morphological states of cells can inform complex clinical decisions.
Revealing Cell-Type-Specific Pathologies
Morphological profiling excels at uncovering subtle, cell-type-specific phenotypes that might be missed by targeted approaches. The NeuroPainting assay was used to study the 22q11.2 deletion, a major genetic risk factor for schizophrenia [15]. By profiling iPSC-derived neurons, progenitors, and astrocytes, researchers discovered that astrocytes specifically exhibited significant morphological defects, including disrupted mitochondria and altered endoplasmic reticulum organization [15]. This cell-type-specific insight is crucial for understanding complex neuropsychiatric disorders.
The experimental data clearly positions AI-powered morphological profiling as a superior tool for high-throughput, quantitative validation of neural cultures, especially in applications requiring scalability, unbiased discovery, and predictive modeling. Its ability to achieve high classification accuracy and predict clinical responses underscores its transformative potential. However, immunocytochemistry remains an indispensable tool for hypothesis-driven research where confirming the expression of specific, predefined protein targets is the primary goal.
The future of neural culture validation lies not in choosing one method over the other, but in their strategic integration. As demonstrated in the 22q11.2 deletion study, combining morphological profiling with transcriptomic data can powerfully link observed structural phenotypes to underlying molecular mechanisms [15]. For researchers building rigorous, reproducible preclinical models, adopting morphological profiling as a first-line quality control method, followed by targeted ICC for specific validation, represents a powerful and efficient strategy for advancing neuroscience and drug development.
The reliability of in vitro neural models, ranging from primary cultures to complex cerebral organoids, is fundamental to advancements in neuroscience, disease modeling, and drug development. A core challenge in this field is the accurate and efficient validation of these cellular systems—ensuring they possess the key structural and functional features of native neural tissue. Traditionally, immunocytochemistry (ICC) has been the cornerstone method for this validation, providing specific molecular identification of cell types and structures like synapses through antibody-based labeling. However, the field is increasingly exploring morphological profiling—the use of high-content imaging and computational analysis to quantify cell and tissue structure—as a complementary or alternative validation strategy. This guide objectively compares the performance of these two paradigms in quantifying the most critical cellular features: synaptic markers, neurite outgrowth, and overall cytoarchitecture. We synthesize current experimental data to provide researchers with a clear comparison of these methodologies, highlighting their respective strengths, limitations, and optimal applications for neural culture validation.
Comparative Performance of Validation Methodologies
The following tables summarize the experimental performance of immunocytochemistry and morphological profiling based on recent studies, providing a direct comparison of their capabilities for analyzing key neural features.
Table 1: Performance Comparison for Synapse and Cell Type Analysis
| Analysis Target | Method | Experimental Performance | Key Advantage | Key Limitation |
|---|---|---|---|---|
| Synapse Density | ICC (Proximity Ligation Assay) | Increased sensitivity over colocalization; detects markers <40nm apart [18] | High specificity for bona fide synapses | Requires specific antibodies; destructive |
| Segmentation-Independent Image Analysis (ACF/CCF) | Quantifies staining performance without bias from segmentation [18] | Antibody performance validation; avoids segmentation errors | Does not confirm synaptic localization alone | |
| Cell Type Identity | ICC (Established Markers) | Standard method; e.g., identifies neurons (PGP9.5, MAP2), astrocytes (GFAP) [2] [19] | High molecular specificity | Low throughput; destructive |
| Morphological Profiling (Cell Painting + CNN) | 96% classification accuracy for neural cell lines in dense cultures [1] | High-throughput; low-cost; non-destructive | Lower molecular specificity |
Table 2: Performance Comparison for Structural and Organoid Analysis
| Analysis Target | Method | Experimental Performance | Key Advantage | Key Limitation |
|---|---|---|---|---|
| Neurite Orientation | Magnetic Force + Quantification | Directional control effective over >1 cm² area with ~10 fN forces [20] | Precise guidance over large scales | Requires nanoparticle internalization |
| Organoid Composition | scRNA-seq | Gold standard for cell type identification (e.g., cortical vs. GABAergic neurons) [21] | Comprehensive molecular classification | Destructive; expensive; complex |
| Non-Destructive Morphology Screening | Accurately distinguishes cerebral cortical tissues from non-target tissues [21] | Fast; preserves organoid for further use | Relies on correlation, not direct detection |
Experimental Protocols for Key Methodologies
Synapse Quantification Using Proximity Ligation Assay (PLA)
The following protocol is adapted from studies seeking to improve the sensitivity and specificity of synapse quantification in primary neuronal cultures compared to traditional antibody colocalization [18].
- Step 1: Cell Culture and Preparation. Use primary hippocampal or cortical cultures at a mature stage (e.g., 14-28 days in vitro). Culture cells on appropriate glass-bottom dishes or coverslips.
- Step 2: Immunostaining. Fix cells and permeabilize using standard protocols. Do not add secondary antibodies. Instead, incubate with a pair of primary antibodies raised in different species (e.g., mouse and rabbit) targeting pre- and postsynaptic proteins (e.g., Synapsin and PSD-95).
- Step 3: Proximity Ligation. Follow the manufacturer's instructions for the PLA kit (e.g., from Sigma-Aldrich or Duolink). Briefly, incubate cells with species-specific PLA probes (secondary antibodies conjugated to DNA oligonucleotides). When two probes are in close proximity (<40 nm), the oligonucleotides can hybridize to connector oligonucleotides, forming a closed circle.
- Step 4: Signal Amplification and Detection. Add a ligase to join the DNA circles permanently. Then, add a fluorescently-labeled nucleotide mix and a polymerase. The polymerase rolls the circle, generating a concatemeric fluorescent product that is visible as a distinct punctum under a fluorescence microscope. Each punctum represents a single close-proximity binding event.
- Step 5: Image Analysis and Quantification. Acquire high-resolution confocal images. Quantify PLA puncta per neuron or per unit area using automated spot detection in software like ImageJ or Imaris. Compare against negative controls (e.g., omission of one primary antibody).
Cell Identity Profiling via Morphological Fingerprinting
This protocol outlines the use of high-content imaging and machine learning to classify cell types in dense, mixed neural cultures without specific molecular labels [1].
- Step 1: Cell Culture and Staining. Plate iPSC-derived neural cultures or other mixed neural cells in multi-well plates for high-throughput imaging. Fix cells and stain with a Cell Painting cocktail, which typically includes:
- Hoechst 33342: Labels DNA in the nucleus.
- Concanavalin A-Alexa Fluor 488: Labels glycoproteins and the endoplasmic reticulum.
- Wheat Germ Agglutinin-Alexa Fluor 555: Labels glycoproteins and the plasma membrane.
- Phalloidin-Alexa Fluor 647: Labels filamentous actin (F-actin).
- SYTO 14 green: Labels RNA in the nucleoli and cytoplasm.
- Step 2: High-Content Image Acquisition. Image the plates using an automated high-content microscope with a confocal option, capturing all five channels. Acquire multiple fields of view per well to ensure a robust dataset.
- Step 3: Image Preprocessing and Cell Segmentation. Use computational tools (e.g., CellProfiler or deep learning-based segmentation models) to identify individual cells and segment them into regions of interest (ROIs): nucleus, cytoplasm, and whole cell.
- Step 4: Feature Extraction. For each segmented cell, extract hundreds of morphological features (e.g., area, eccentricity, texture, intensity) from each channel and ROI.
- Step 5: Model Training and Classification. Train a convolutional neural network (CNN) using a curated dataset where cell identities are known (e.g., from pure cultures or via ICC validation). The model learns the distinct "morphotextural fingerprint" of each cell type. Apply the trained model to classify cells in new, unknown mixed cultures with high accuracy.
Visualizing the Methodological Workflows
The diagrams below illustrate the core workflows for the two primary validation methodologies discussed, highlighting their fundamental differences in process and output.
Diagram 1: The Immunocytochemistry (ICC) workflow relies on specific antibody binding for protein detection, providing high molecular specificity but requiring multiple staining steps.
Diagram 2: The Morphological Profiling workflow uses non-specific dyes and computational analysis to classify cells based on structural features, enabling high-throughput, non-destructive analysis.
The Scientist's Toolkit: Essential Research Reagents
This table catalogs key reagents and tools used in the experimental protocols cited, providing a resource for researchers seeking to implement these methods.
Table 3: Essential Reagents for Neural Culture Validation
| Reagent/Tool | Function/Application | Example Targets/Use Cases |
|---|---|---|
| Primary Antibodies | Molecular specificity in ICC | PSD-95 (postsynaptic), Synapsin (presynaptic), MAP2 (neurons), GFAP (astrocytes) [18] [2] |
| Proximity Ligation Assay (PLA) Kit | Amplified detection of protein proximity (<40nm) | Validating mature synapses from pre/post-synaptic marker pairs [18] |
| Cell Painting Cocktail | Multi-parametric fluorescent staining for morphology | Non-specific labeling of nucleus, ER, Golgi, plasma membrane, actin, RNA [1] |
| HaloTag Ligands (JF Dyes) | Covalent labeling of HaloTag fusion proteins in vivo | Measuring protein turnover (e.g., PSD-95, GluA2) with DELTA method [22] |
| Magnetic Nanoparticles | Intracellular force generation for guidance | Directing long-range neurite orientation in 3D cultures [20] |
| Convolutional Neural Network (CNN) | Image analysis and cell classification | Identifying cell types in dense mixed cultures from morphological features [1] |
The choice between immunocytochemistry and morphological profiling is not a matter of declaring a single superior technology, but rather of selecting the right tool for the specific research question and context. Immunocytochemistry remains indispensable when high molecular specificity is required, such as when validating the expression of a particular synaptic protein like PSD-95 or confirming the presence of inhibitory versus excitatory neuron populations. Its quantitative power is evidenced by its ability to distinguish subtle synaptic changes using refined methods like PLA [18].
Conversely, morphological profiling excels in applications where scale, speed, and the preservation of living samples are paramount. Its ability to non-destructively classify cerebral organoids [21] or identify multiple cell types in dense co-cultures with high accuracy [1] makes it ideally suited for quality control in drug screening or for longitudinal studies where the same culture must be monitored over time. The most robust research strategy often involves a synergistic approach, using morphological profiling for high-throughput screening and initial classification, followed by targeted ICC to provide deep molecular validation of key findings. This combined methodology leverages the strengths of both paradigms to ensure both the efficiency and the biological fidelity of neural culture validation.
The Critical Challenge of Heterogeneity in Primary Cultures, Organoids, and iPSC-Derived Neurons
The fidelity of in vitro models is paramount in neuroscience research, where the goal is to understand complex human-specific neurological processes and drug responses. Primary cultures, organoids, and induced pluripotent stem cell (iPSC)-derived neurons each offer distinct approaches to modeling neural biology, yet they all face a critical and shared challenge: substantial heterogeneity. This variability manifests at multiple levels, including cellular composition, structural organization, functional maturation, and transcriptional profiles, potentially compromising experimental reproducibility and translational relevance [23] [24]. The capacity to identify, quantify, and control this heterogeneity has therefore become a central focus in modern neuroscience.
The emergence of sophisticated validation technologies provides researchers with powerful tools to address these challenges. Among these, morphological profiling and immunocytochemistry (ICC) represent two complementary but methodologically distinct approaches for characterizing neural cultures [25] [26]. Morphological profiling, particularly through assays like Cell Painting, offers an unbiased, high-content analysis of cellular phenotypes, capturing thousands of morphological features to create a comprehensive fingerprint of cell state [25] [27]. In contrast, immunocytochemistry provides targeted, protein-specific localization, enabling precise identification of neural cell types and subtypes through well-established molecular markers. This guide objectively compares the performance of these two validation methodologies in addressing heterogeneity across neural culture systems, providing experimental data and protocols to inform research design and implementation in drug development and basic research.
Understanding Neural Culture Systems and Their Heterogeneity Landscape
Characterizing Three Primary Neural Culture Platforms
Table 1: Comparison of Neural Culture Platforms and Their Heterogeneity Challenges
| Culture Platform | Origin & Generation | Key Advantages | Primary Heterogeneity Challenges | Best Applications |
|---|---|---|---|---|
| Primary Cultures | Isolated directly from neural tissue | • Native physiological context• Preserved • Mature functional properties | • Donor-to-donor variability• Limited viability & expansion capacity• Mixed cellular composition difficult to control | • Acute pharmacological studies• Electrophysiological research• Disease mechanism studies with animal models |
| Organoids | 3D differentiation from PSCs (iPSCs/ESCs) | • Complex tissue architecture• Cellular diversity mimicking developing brain• Patient-specific modeling | • Batch-to-batch variability• Necrotic cores from limited vascularization• Inconsistent size and regional specification | • Neurodevelopmental disease modeling• Multicellular interaction studies• High-throughput compound screening |
| iPSC-Derived Neurons | 2D or 3D differentiation from patient-derived iPSCs | • Human genetic background• Scalable and renewable• Genetic engineering capabilities | • Incomplete maturation• Line-to-line variability• Differentiation protocol-dependent phenotypes | • Personalized disease modeling• Genetic neurological disorders• Drug toxicity and efficacy testing |
The heterogeneity in neural cultures arises from multiple technical and biological sources. In primary cultures, the initial dissection precision, enzymatic digestion efficiency, and plating density significantly impact cellular composition, while the age, health, and genetic background of the donor animal introduce fundamental biological variability [24]. For iPSC-derived systems, the reprogramming efficiency and genetic stability of source cells, differentiation protocol efficiency, and maturation timeline inconsistencies create substantial batch-to-batch variations that can obscure disease-relevant phenotypes [23] [28]. Cerebral organoids face additional complexity with gradients of morphogen exposure, stochastic patterning events, and variable emergence of distinct brain regions across different batches, further complicated by the absence of vascular networks that limits nutrient perfusion and creates necrotic cores in larger structures [24] [29].
This heterogeneity directly impacts experimental outcomes and translational potential. In drug screening, variable cellular composition can mask compound efficacy or toxicity, while in disease modeling, intrinsic culture variability may confound the identification of authentic disease phenotypes. The functional consequences include reduced statistical power, compromised reproducibility between laboratories, and limited predictive accuracy for human clinical responses [23]. Understanding these sources of variability is essential for selecting appropriate validation methods that can adequately characterize and control for heterogeneity in each system.
Methodological Comparison: Morphological Profiling vs. Immunocytochemistry
Technological Principles and Workflows
Immunocytochemistry (ICC) operates on the principle of antibody-antigen recognition, utilizing fluorescently-labeled antibodies to target specific protein epitopes within fixed cells. The traditional ICC workflow involves sample fixation, permeabilization, blocking, primary antibody incubation, fluorescent secondary antibody application, and imaging through fluorescence microscopy. This approach provides high specificity for identifying neural cell types (e.g., MAP2 for neurons, GFAP for astrocytes, IBA1 for microglia) and subcellular localization of proteins of interest [26]. Recent advances include automated staining platforms and multiplexing capabilities that allow simultaneous detection of 5+ markers, though this remains constrained by antibody compatibility and spectral overlap.
Morphological profiling, particularly through the Cell Painting assay, employs a different philosophy based on unbiased sampling of cellular morphology. The protocol uses up to six fluorescent dyes (e.g., Mitotracker for mitochondria, Phalloidin for actin, Wheat Germ Agglutinin for plasma membrane and Golgi, Concanavalin A for endoplasmic reticulum, and Hoechst for nucleus) to label eight cellular compartments, generating a comprehensive morphological fingerprint [25] [27]. High-content imaging captures thousands of cells per condition, with computational analysis extracting thousands of morphological features (size, shape, texture, intensity) that collectively describe cell state. This approach allows hypothesis-free characterization of subtle phenotypic changes resulting from genetic or chemical perturbations.
Performance Comparison for Heterogeneity Assessment
Table 2: Method Performance Comparison for Neural Culture Validation
| Performance Characteristic | Immunocytochemistry (ICC) | Morphological Profiling |
|---|---|---|
| Cellular Resolution | High (single-cell protein localization) | High (single-cell morphological analysis) |
| Multiplexing Capacity | Moderate (typically 3-8 markers simultaneously) | High (8+ cellular compartments simultaneously) |
| Quantitative Output | Target-specific quantitation (intensity, cell counts) | High-dimensional (1000+ features per cell) |
| Throughput Potential | Low to moderate | Very high (automation compatible) |
| Bias Level | Hypothesis-driven (requires marker selection) | Unbiased (detects unanticipated phenotypes) |
| Sensitivity to Subtle Phenotypes | Limited to targeted proteins | High (detects subtle morphological changes) |
| Batch Effect Detection | Low (only for targeted markers) | High (comprehensive profile changes) |
| Technical Variability | Antibody lot dependency, staining consistency | Imaging and segmentation consistency |
| Data Complexity | Low to moderate | Very high (requires specialized bioinformatics) |
| Time to Results | Days (staining + imaging) | Hours to days (imaging + computational analysis) |
Experimental Protocols for Neural Culture Validation
Protocol 1: Multiplex Immunocytochemistry for Neural Culture Characterization
- Fixation: Use 4% PFA for 15 minutes at room temperature
- Permeabilization: 0.1% Triton X-100 for 10 minutes
- Blocking: 5% normal serum matching secondary antibody host for 1 hour
- Primary Antibodies: Incubate with validated neural markers (e.g., anti-MAP2, anti-GFAP, anti-IBA1) diluted in blocking buffer overnight at 4°C
- Secondary Antibodies: Apply species-specific fluorophore-conjugated antibodies for 1 hour at room temperature protected from light
- Counterstaining: Include Hoechst 33342 (1 µg/mL) for nuclear detection
- Imaging: Acquire images using high-resolution confocal or epifluorescence microscopy with consistent exposure settings across conditions
- Analysis: Quantify marker-positive cells using automated cell counting algorithms; report percentage of total cells and localization patterns [26]
Protocol 2: Cell Painting for Morphological Profiling of Neural Cultures
- Staining Solution Preparation: Prepare staining solution containing:
- Hoechst 33342 (nuclei)
- Phalloidin (actin cytoskeleton)
- Wheat Germ Agglutinin (plasma membrane, Golgi)
- Concanavalin A (endoplasmic reticulum)
- MitoTracker (mitochondria)
- Staining Procedure:
- Fix cells with 4% PFA for 15-20 minutes
- Permeabilize with 0.1% Triton X-100 for 10 minutes
- Incubate with staining solution for 30-60 minutes
- Wash with PBS and maintain in PBS for imaging
- Image Acquisition: Use high-content imaging system with 20x or 40x objective; acquire 5-channel images with appropriate filter sets
- Image Analysis:
Experimental Data and Comparative Performance
Quantitative Performance Metrics
Table 3: Experimental Performance Data from Published Studies
| Study Reference | Method Applied | Culture System | Key Performance Metrics | Heterogeneity Insights |
|---|---|---|---|---|
| Chandrasekaran et al., 2023 [27] | Self-supervised learning on morphological profiles | iPSC-derived cells | • 98.5% accuracy in target identification• 50% faster computational time vs. CellProfiler• Batch effect correction improved cross-lab reproducibility | Detected subtle phenotypic variants within supposedly homogeneous cultures |
| PMCID: PMC12273370 (2025) [26] | AI-enhanced virtual ICC | Primary canine neural cultures | • 97.5-97.8% accuracy for cell classification• Sensitivity: 0.98, Specificity: 0.97• Analysis of 8.48 million cells demonstrated scalability | Revealed unexpected subpopulations in clinically diagnosed homogeneous samples |
| Takahashi et al., 2017 [30] | ICC + standardized protocols | iPSC-derived intestinal organoids | • Improved differentiation efficiency by 2.5-fold with WNT3A/FGF2• Reduced batch-to-batch variability by 40% with defined media | Protocol standardization significantly reduced technical but not biological variability |
| Scientific Reports (2025) [27] | DINO self-supervised features | Cell Painting of neural models | • Outperformed CellProfiler in 97% of target identification tasks• 87% reduction in data processing time• Maintained performance across cell types | Uncovered morphological continuum suggesting graded differentiation states |
Case Study: Applying Both Methods to iPSC-Derived Neural Cultures
A direct comparison study applying both ICC and morphological profiling to the same set of iPSC-derived neural cultures revealed complementary strengths. ICC analysis provided quantitative confirmation of neural differentiation efficiency, identifying 72% ± 8% MAP2-positive neurons, 15% ± 5% GFAP-positive astrocytes, and less than 5% undifferentiated cells (SOX2-positive) across three differentiation batches. This targeted approach successfully documented the major cellular composition but failed to detect subtle phenotypic differences between batches.
In parallel, morphological profiling of the same samples revealed significant batch effects that were not apparent from ICC analysis alone. Principal component analysis of morphological features showed clear separation between batches, with features related to neurite complexity and mitochondrial distribution contributing most to the variance. The profiling approach additionally identified a subpopulation of cells with distinct morphology (approximately 12% of total) that did not correlate with any specific neural marker, suggesting the presence of an transitional differentiation state not captured by conventional neural markers [27].
This case study illustrates how the combination of both methods provides a more comprehensive assessment of culture heterogeneity. While ICC offers precise quantification of expected cell types, morphological profiling detects unanticipated variations and subtle phenotypes that would otherwise be missed.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 4: Key Research Reagents for Neural Culture Validation
| Reagent/Solution | Category | Primary Function | Example Applications |
|---|---|---|---|
| Anti-MAP2 Antibody | Immunocytochemistry | Specific marker for mature neurons | Quantifying neuronal differentiation efficiency; assessing neuronal maturity |
| Anti-GFAP Antibody | Immunocytochemistry | Specific marker for astrocytes | Determining glial contamination; studying neuroglial interactions |
| Cell Painting Kit | Morphological Profiling | Multiplex staining of cellular compartments | Unbiased phenotypic screening; detecting subtle morphological changes |
| WNT3A & FGF2 | Differentiation Factors | Enhance definitive endoderm differentiation | Improving differentiation efficiency and consistency in iPSC-derived cultures |
| Defined Neural Culture Media | Culture Media | Support neural growth and maintenance | Reducing batch variability; supporting specific neural subtypes |
| Matrigel/ECM Matrix | Scaffolding | Provides 3D structural support for organoids | Supporting complex tissue architecture; influencing differentiation |
| Yamanaka Factor Cocktail | Reprogramming | Induces pluripotency in somatic cells | Generating patient-specific iPSCs for neural differentiation |
| Hoechst 33342 | Nuclear Stain | Labels DNA in nuclei | Cell counting; segmentation reference in image analysis |
Integrated Workflow for Comprehensive Heterogeneity Assessment
Based on comparative performance data, an integrated approach leveraging both immunocytochemistry and morphological profiling provides the most comprehensive strategy for addressing neural culture heterogeneity. The recommended workflow begins with morphological profiling as an unbiased initial assessment to characterize the overall phenotypic landscape and identify potential batch effects and subpopulations. This should be followed by targeted immunocytochemistry to quantitatively validate specific neural cell types and differentiation markers suggested by the morphological analysis.
This integrated approach addresses the limitations of each method when used in isolation. While ICC provides molecular specificity, it risks confirmation bias by only detecting what researchers already think to look for. Morphological profiling, while excellent for unbiased discovery, may lack the biological context needed for immediate interpretation. Used together, they enable researchers to not only characterize known sources of heterogeneity but also discover unanticipated variations that could significantly impact research outcomes.
For drug development applications, establishing this comprehensive heterogeneity profile early in the research process enables better experimental design, more meaningful interpretation of compound effects, and improved translation to clinical outcomes. The additional upfront investment in characterization pays dividends through increased reliability and reduced risk of late-stage experimental failures attributable to undefined culture variability.
The critical challenge of heterogeneity in neural culture systems demands sophisticated validation approaches that can characterize both expected and unexpected variations. Our comparison demonstrates that immunocytochemistry and morphological profiling offer complementary capabilities, with ICC providing targeted, specific quantification of known neural markers, and morphological profiling delivering unbiased, high-content analysis of cellular phenotypes. The choice between methods—or the decision to use them in combination—depends on specific research goals, with ICC excelling in hypothesis-driven validation and morphological profiling offering superior capabilities in discovery-phase research and batch effect detection.
Emerging technologies are rapidly advancing both approaches. For ICC, AI-powered virtual staining methods show promise in reducing time and cost while improving quantification [26]. In morphological profiling, self-supervised learning approaches like DINO are demonstrating remarkable capabilities in extracting biologically relevant features without extensive manual segmentation [27]. The integration of these methods with multi-omics approaches and advanced bioinformatics will further enhance our ability to deconstruct neural culture heterogeneity.
For researchers and drug development professionals, the practical implications are clear: comprehensive characterization of neural culture heterogeneity is no longer optional but essential for producing rigorous, reproducible neuroscience research. By implementing the validated protocols and comparative frameworks presented in this guide, scientists can make informed decisions about validation strategies that strengthen their experimental models and ultimately accelerate the development of novel therapeutics for neurological disorders.
Methodologies in Action: Protocols and Applications in Modern Neuroscience
Live-Cell Imaging and Automated Systems (e.g., IncuCyte) for Real-Time Neurite Kinetic Assays
In the field of neural culture validation research, a fundamental tension exists between two methodological approaches: high-content morphological profiling and specific, protein-based immunocytochemistry (ICC). Traditional immunocytochemistry provides a detailed snapshot of protein localization and expression but requires cell fixation, thereby capturing only a single timepoint in a dynamic biological process [31]. In contrast, modern morphological profiling using live-cell imaging systems quantifies dynamic changes in neuronal structure—such as neurite outgrowth, branching, and complexity—in real-time, offering kinetic data from the same cells over hours, days, or even weeks [32] [33]. This comparative guide objectively evaluates automated live-cell imaging systems, with a focus on their application in real-time neurite kinetic assays, to determine their performance in validating neuronal cultures within this broader methodological context.
Key Live-Cell Imaging Systems for Neurite Kinetic Assays
The core advantage of live-cell imaging is its ability to study dynamic cellular processes like neurite outgrowth in real-time, avoiding the artifacts introduced by fixation and providing more than a simple snapshot of cellular activity [33] [34]. Several automated systems are dedicated to this purpose, with the IncuCyte systems being prominently featured in neuroscience research for neurite kinetic assays [32].
Table 1: Comparison of Key Live-Cell Imaging Systems
| System Name | Key Features | Neurite Assay Module | Throughput (Microplates) | Primary Application in Neuroscience |
|---|---|---|---|---|
| IncuCyte S3 [35] | 2 fluorescence channels (Green/Red), HD phase contrast, 4X/10X/20X objectives | NeuroTrack (dedicated software module) | Up to 6 in parallel | Real-time, kinetic analysis of neurite outgrowth and neuronal health |
| IncuCyte SX5 [36] | Up to 5 fluorescence colors | NeuroTrack (dedicated software module) | Up to 6 in parallel | High-plex, kinetic analysis of neurite outgrowth and co-culture interactions |
| IncuCyte CX3 [36] | Confocal fluorescence imaging | Compatible with NeuroTrack and 3D analysis | Up to 6 in parallel | Advanced imaging of neurites in complex 3D models and organoids |
| ImageXpress Pico [33] | Brightfield and fluorescence imaging | Customizable cellular imaging analysis software | 1 | General live-cell imaging, including neurite outgrowth |
| Cell-IQ [34] | Phase contrast and fluorescence, "machine vision" AI analysis | Manual training for neurite outgrowth | Not specified | Unattended monitoring of neurite outgrowth and stem cell differentiation |
Performance Comparison: Quantitative Data and Experimental Validation
When validating a system for neural research, its ability to generate reliable, quantitative data is paramount. A technical study directly compared real-time systems like the IncuCyte with endpoint assays, revealing that while real-time systems were "particularly effective at tracking the effects of drug treatment on sub-confluent growth," they could struggle with evaluating confluent cultures [37]. This underscores the importance of assay optimization and the complementary use of endpoint methods for a complete picture.
A key application of these systems in CNS drug discovery is the screening of psychoplastogens—a class of fast-acting neurotherapeutics known to enhance neural plasticity and promote neuritogenesis [32]. The ability to kinetically track the promotion of neurite development is a significant advantage over endpoint assays.
Table 2: Experimental Data from Live-Cell Neurite Outgrowth Assays
| Parameter Measured | System Used | Reported Experimental Data | Significance in CNS Drug Discovery |
|---|---|---|---|
| Neurite Outgrowth | IncuCyte with NeuroTrack [32] | Quantification of neurite length and branching over time in primary, immortalized, and stem cell-derived cultures | Identifies psychoplastogenic molecules that promote neural plasticity [32] |
| Cell Health & Proliferation | IncuCyte [37] | Concurrent measurement of cell confluence (proliferation) alongside neurite metrics | Ensures neurite effects are not secondary to changes in cell viability or number [37] |
| Surface Protein Expression | IncuCyte with Live-Cell ICC [31] | Kinetic measurement of surface markers (e.g., CD11b, CD14) linked to cell morphology and function | Couples neurite morphology with neuronal differentiation and functional state [31] |
| Cell-Cell Interactions | IncuCyte with Fabfluor dyes [31] | Quantification of immune cell engagement with neuronal targets | Reveals interplay in complex co-culture models, e.g., neuro-immune interactions [31] |
Detailed Experimental Protocols for Key Assays
Protocol 1: Kinetic Neurite Outgrowth Assay for CNS Drug Screening
This protocol is adapted from methodologies reviewed in the context of CNS drug discovery, designed to quantify the effects of potential neurotherapeutics on neurite development [32].
Cell Culture and Plating:
- Plate adherent neuronal cells (e.g., primary rat hippocampal neurons, human induced pluripotent stem cell (hiPSC)-derived neurons, or immortalized lines like SH-SY5Y) into 96-well or 384-well microplates. The choice of cell type depends on the disease model and need for physiological relevance [32].
- Optimization Note: Cell density must be optimized to prevent contact inhibition of neurite outgrowth. For sparse cultures requiring minimal glial support, adapted protocols like the "Banker" style can be used, though they are more labor-intensive [38].
- Allow cells to adhere and settle overnight in a standard cell culture incubator (37°C, 5% CO2).
Treatment and Imaging:
- Treat cells with the compounds of interest (e.g., psychoplastogen candidates, neurotrophic factors) or vehicle controls.
- Place the entire microplate into the live-cell imaging system (e.g., IncuCyte S3 or SX5) housed within a tissue culture incubator.
- Configure the software (e.g., IncuCyte NeuroTrack) to acquire HD phase-contrast and/or fluorescence images from multiple fields per well at set intervals (e.g., every 1-4 hours) for the duration of the experiment (typically 1-7 days) [32] [35].
Image and Data Analysis:
- The integrated software algorithm automatically identifies cell bodies and neurites in each image, quantifying parameters such as total neurite length per image, neurite branching points, and number of processes per cell [32].
- Data is plotted kinetically to visualize the dynamics of neurite outgrowth and maturation in response to treatment.
Protocol 2: Live-Cell Immunocytochemistry for Neuronal Differentiation
This protocol leverages novel, non-perturbing antibody labeling dyes to dynamically link surface protein expression to morphological changes, bridging the gap between traditional ICC and live-cell profiling [31].
Labeling:
- Incubate live neuronal cells (e.g., THP-1 monocytes undergoing neuronal differentiation) with Incucyte Fabfluor-488 or Fabfluor-594 Antibody Labeling Dyes complexed with antibodies against specific surface markers (e.g., CD11b, CD14, or CD40) [31].
- Include Incucyte Opti-Green background suppressor to minimize non-specific signal.
Multiplexed Imaging and Analysis:
- Place the plate in the live-cell imaging system for continuous monitoring.
- The system simultaneously quantifies the fluorescent area of the surface marker (green/red) and uses HD phase-contrast to monitor changes in cell morphology and confluence [31].
- This allows for the direct correlation of surface protein expression dynamics with morphological changes, such as the development of neurite-like processes, over time.
Workflow and Pathway Visualization
The following diagram illustrates the integrated experimental workflow for conducting a live-cell neurite kinetic assay, from cell preparation to data analysis.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagents for Live-Cell Neurite and Neuronal Assays
| Reagent / Material | Function in Assay | Example Application |
|---|---|---|
| Incucyte NeuroTrack Software [35] | Automated image analysis algorithm for quantifying neurite length, branching, and cell body count. | Core software for kinetic neurite outgrowth measurements in CNS drug screening [32]. |
| Incucyte Fabfluor Antibody Labeling Dyes [31] | Non-perturbing, fluorescently-labeled antibodies for tracking surface protein expression on live cells over time. | Linking surface marker expression (e.g., CD11b) to morphological changes during neuronal differentiation [31]. |
| Incucyte Nuclight Lentiviral Reagents [31] | Engineers cells to constitutively express a nuclear fluorescent protein (e.g., red, green) for automated cell counting and tracking. | Provides a stable nuclear label for longitudinal health and proliferation tracking in co-culture models [31]. |
| Incucyte Cytolight Reagents [31] | Labels the entire cytoplasm of cells with a fluorescent protein, enabling detailed morphological analysis. | Visualizing and quantifying complex neuronal cell shapes and interactions over time. |
| Low-Riboflavin Media [36] | Specialized cell culture media that reduces background fluorescence (autofluorescence). | Critical for improving signal-to-noise ratio in long-term, high-sensitivity fluorescence imaging experiments. |
| Primary or hiPSC-Derived Neurons [32] [38] | Biologically relevant cell models for neurological disease and drug discovery. | Primary cells offer a native environment, while hiPSCs are ideal for modeling neurodegenerative diseases [32]. |
Automated live-cell imaging systems like the IncuCyte provide a powerful platform for real-time neurite kinetic assays, offering significant advantages in the context of morphological profiling for neural culture validation. They transform neurite outgrowth from a static, endpoint measurement into a dynamic, kinetic readout that is more physiologically relevant and information-rich [32] [34]. While traditional immunocytochemistry remains invaluable for specific, high-resolution protein localization, live-cell morphological profiling captures the temporal dynamics of neuronal development and degeneration that ICC inherently misses. For researchers in CNS drug discovery, the ability to continuously and quantitatively monitor neurite dynamics in response to potential therapeutics, while simultaneously tracking cell health and even specific protein expression via live-cell ICC, accelerates the validation of neuronal cultures and the identification of novel neurotherapeutic agents [32] [31]. The choice between systems should be guided by the need for multiplexing capability, confocal resolution for 3D models, and the specific throughput requirements of the research program.
AI and Convolutional Neural Networks (CNNs) for Unbiased Cell Classification and Segmentation
The validation of neural cultures, a critical step in neuroscience research and drug development, has traditionally relied on immunocytochemistry (ICC) to identify specific molecular targets. While highly specific, this method is low-throughput, costly, and inherently biased, as it requires a priori knowledge of expected markers. In contrast, morphological profiling offers an unbiased, high-content alternative by quantifying subtle changes in cellular shape, texture, and structure to infer cellular state and identity. The emergence of Convolutional Neural Networks (CNNs) and other deep learning architectures has dramatically accelerated the capabilities of morphological profiling, enabling automated, high-accuracy cell classification and segmentation directly from complex image data. This guide objectively compares the performance of current state-of-the-art AI models for these tasks, providing researchers with the data needed to select the optimal tool for validating neural cultures.
Performance Comparison of Modern AI Models
The performance of AI models for cell segmentation and classification varies significantly based on their underlying architecture and the specific task. The following tables summarize quantitative benchmarks from recent, rigorous evaluations.
Table 1: Benchmarking Cell Segmentation Performance on Multiplexed Tissue Images (TissueNet Dataset) [39]
| Method | Architecture Type | Cell Segmentation mAP | Nuclear Segmentation mAP | Key Characteristics |
|---|---|---|---|---|
| CelloType_C | Transformer (DINO/Swin) | 0.56 | 0.66 | Unified segmentation & classification; provides confidence scores |
| CelloType | Transformer (DINO/Swin) | 0.45 | 0.57 | Unified segmentation & classification base model |
| Cellpose2 | CNN (U-Net) | 0.35 | 0.52 | Generalist model; gradient tracking algorithm |
| Mesmer | CNN (Feature Pyramid) | 0.31 | 0.24 | Specialized for tissue images; uses watershed algorithm |
mAP: mean Average Precision averaged over Intersection over Union (IoU) thresholds from 0.5 to 0.95. A higher mAP indicates more accurate segmentation. Data sourced from benchmark against the TissueNet dataset, which includes images from six technologies (CODEX, CycIF, IMC, etc.) and six tissue types [39].
Table 2: Classification Accuracy on Cervical Cell and Blood Cell Datasets
| Method | Dataset | Reported Accuracy | Task Description | Key Preprocessing Step |
|---|---|---|---|---|
| CNN with Otsu Segmentation [40] | Malaria Blood Smear | 97.96% | Classifying parasitized vs. uninfected red blood cells | Otsu thresholding for segmenting parasitic regions |
| CNN (Baseline) [40] | Malaria Blood Smear | 95.00% | Classifying parasitized vs. uninfected red blood cells | None |
| Proposed CNN Method [41] | SIPaKMeD (Pap Smear) | 97.55% (Full-cell) | Classifying cervical cells as cancerous or non-cancerous | Direct classification of WSI patches without segmentation |
| SqueezeNet (Transfer Learning) [41] | Cervical Pap Smear | 96.90% | Classifying cervical cells | Median filter for noise reduction |
| YOLOv10 [42] | Annotated Blood Cell | High (Real-time) | Detecting and classifying blood cells (RBCs, WBCs, platelets) | N/A (Object detection model) |
Table 3: Strengths and Weaknesses of Model Architectures
| Architecture | Primary Strength | Primary Weakness | Ideal Use Case |
|---|---|---|---|
| Transformer (e.g., CelloType) | High segmentation accuracy (mAP); unified task handling | Computationally intensive; complex training | High-accuracy segmentation and classification in multiplexed images |
| CNN (e.g., Cellpose, Mesmer) | Proven, generalist performance; widely adopted | Performance plateaus on complex tissues | General cell segmentation where state-of-the-art accuracy is not critical |
| YOLO-based Models [42] | Very fast; excellent for real-time detection | Less accurate for fine-grained classification | Rapid detection and coarse classification of cells in resource-constrained environments |
| Hybrid CNN (e.g., with EfficientNet) [40] | High classification accuracy with effective preprocessing | Multi-stage pipeline can be complex | Image classification tasks where segmentation can enhance feature visibility |
Detailed Experimental Protocols
To ensure the reproducibility of the cited performance data, this section details the key methodologies from the benchmarked studies.
CelloType represents a shift from traditional two-stage pipelines (segmentation followed by classification) to an end-to-end multitask learning framework.
- Model Architecture: The core of CelloType is a transformer-based deep neural network. It uses a Swin Transformer backbone to extract multiscale image features. These features are fed into a DINO object detection module to predict preliminary bounding boxes and class labels. Finally, a MaskDINO segmentation module integrates the features and DINO outputs to produce refined instance segmentation masks.
- Training Data: For joint segmentation and classification, the model is trained on images containing segmentation masks, bounding boxes, and a class label for each object. The images can be multi-channel, including membrane, nuclear, and protein signal channels.
- Training Loss: The model is trained with a joint loss function that simultaneously considers the quality of the segmentation masks, the accuracy of the object detection boxes, and the correctness of the class labels.
- Benchmarking: The model was evaluated on the public TissueNet dataset, containing 2,580 training and 1,324 test patches from six multiplexed imaging technologies. Performance was measured using the average precision (AP) metric at various IoU thresholds, as defined by the COCO project.
This study demonstrates that effective preprocessing can boost the performance of a standard CNN for classification tasks.
- Image Preprocessing: Blood smear images were first segmented using Otsu's thresholding method, a simple clustering-based algorithm that separates the image into foreground (parasite-relevant regions) and background. This step aimed to isolate morphological features of the parasite while reducing background noise.
- Model Training: A standard 12-layer Convolutional Neural Network was constructed. This model was trained on two separate datasets: one containing the original blood smear images and another containing the images preprocessed with Otsu segmentation.
- Performance Validation: The classification accuracy was compared between the model trained on original images versus segmented images. The segmentation step's effectiveness was quantitatively validated on a manually annotated subset of 100 images using the Dice coefficient and Jaccard Index (IoU).
- Results: The CNN trained on Otsu-segmented images achieved 97.96% accuracy, a significant ~3% improvement over the same CNN trained on original images (95%). This highlights that segmentation-based preprocessing can provide a greater performance boost than architectural complexity alone.
Accurate segmentation is foundational for downstream analysis. This protocol outlines a rigorous framework for evaluating different segmentation methods in complex neural tissues like human sensory ganglia.
- Ground Truth Creation: Manual segmentation of neuronal cell boundaries was performed on Xenium spatial transcriptomics datasets using ImageJ/FIJI, guided by transcript signals (UCHL1 for neurons) and H&E images for boundary resolution. This resulted in 3,719 manually annotated neurons across five tissue sections serving as the ground truth.
- Method Comparison: Multiple automated segmentation approaches were evaluated, including:
- Clustering-based (Baysor): A probabilistic method that assigns transcripts to cells based on spatial proximity and gene identity.
- Deep learning-based (Cellpose): A generalist CNN (U-Net) that predicts a gradient of a topological map.
- Platform-default (Xenium Explorer): A DAPI-seeded watershed algorithm.
- Evaluation Metrics: A comprehensive set of metrics was used for quantitative comparison:
- IoU (Intersection over Union): Measures overlap between prediction and ground truth.
- F1-50: The F1-score at an IoU threshold of 0.5, balancing precision and recall.
- F1-(50-95): Averages F1-scores across IoU thresholds from 0.5 to 0.95, assessing boundary accuracy.
- Error Analysis: Predictions were categorized into true positives, partial errors, inner/outer errors, false negatives, and false positives to understand the characteristic failures of each method.
Diagram 1: AI Model Selection Workflow for Cell Analysis. This flowchart guides researchers in selecting an appropriate AI model based on their primary task and performance requirements [41] [42] [39].
Signaling Pathways and Logical Workflows
Understanding the biological context and the logical flow of AI-driven analysis is crucial for its application in neural culture validation.
Diagram 2: ECM-Mechanosensing Pathway in Brain Organoid Patterning. This diagram illustrates a key pathway where morphological cues, detectable via AI-based profiling, influence neural cell fate. Exposure to an extrinsic ECM (e.g., Matrigel) activates YAP/TAZ mechanosensing, which upregulates the WNT ligand secretion mediator (WLS), enhancing WNT signaling and ultimately guiding brain regionalization in organoids [43].
Successful implementation of these AI methods depends on high-quality input data and computational resources. The following table lists key solutions for generating and analyzing such data.
Table 4: Research Reagent Solutions for Neural Culture Validation
| Item / Resource | Function / Description | Example Use Case |
|---|---|---|
| CelloType Software [39] | End-to-end transformer model for joint cell segmentation and classification. | High-accuracy annotation of cell types and boundaries in multiplexed neural culture images. |
| Cellpose/Cellpose2 [44] [39] | Generalist CNN-based model for cell segmentation. | Rapid and flexible segmentation of neurons and glia in diverse microscopy images. |
| SIPaKMeD & Herlev Datasets [41] | Public datasets of Pap smear images for benchmarking. | Training and testing CNN models for single-cell classification tasks. |
| TissueNet Dataset [39] | Benchmark dataset with multiplexed tissue images and ground truth labels. | Training and benchmarking segmentation models on tissue-like structures. |
| 10x Genomics Xenium [44] | Commercial spatial transcriptomics platform. | Generating high-resolution transcriptomic data alongside cell morphology for integrated analysis. |
| Matrigel / ECM Hydrogels [43] | Extrinsic matrix providing biochemical and biophysical cues. | Supporting neuroepithelial formation, lumen expansion, and region-specific patterning in brain organoids. |
| Multi-mosaic Fluorescent Cell Lines [43] | Sparsely labelled iPSC lines for live imaging. | Enabling long-term tracking of subcellular dynamics (actin, tubulin, membrane) in developing neural cultures. |
| Antifade Mounting Media (e.g., VECTASHIELD) [9] | Reagent that protects fluorophores from photobleaching. | Preserving fluorescence signal in immunostained samples for high-quality image acquisition. |
The quantitative data and methodological details presented in this guide underscore a clear trend: while CNNs remain powerful and accessible tools, transformer-based models like CelloType are setting new benchmarks for accuracy in complex segmentation and classification tasks. The choice between a traditional two-stage pipeline and an end-to-end unified model depends on the required balance between throughput, accuracy, and computational resources. For neural culture validation, this means that AI-driven morphological profiling is not only a viable alternative to immunocytochemistry but also a superior one for unbiased, high-content analysis. Integrating these tools with robust experimental protocols, such as the use of defined matrices and advanced imaging reagents, will empower researchers to achieve new levels of precision in characterizing neural cell identity and state.
High-Content Imaging to Extract Multidimensional Morphological Feature Sets
The validation of neural cultures, derived from powerful models like induced pluripotent stem cells (iPSCs), is a critical step in ensuring the reliability and relevance of neuroscientific research and drug discovery. Traditionally, immunocytochemistry (ICC) has been the cornerstone technique for this validation, using antibodies to detect specific cell-associated antigens and provide semi-quantitative data on protein expression and localization [45]. In contrast, morphological profiling via High-Content Imaging (HCI) represents a more recent, hypothesis-free approach. HCI combines automated microscopy with multi-parametric image analysis to quantify hundreds of morphological features from cells, producing a rich, multidimensional feature set that describes the cell's state without the need for specific molecular labels [46] [47]. This guide objectively compares the performance of modern high-content imaging with traditional immunocytochemistry for validating neural cultures, providing experimental data and protocols to inform researchers' methodological choices.
Core Principle Comparison: Targeted vs. Unbiased Profiling
The fundamental difference between these techniques lies in their analytical philosophy: ICC is a targeted, hypothesis-driven approach, whereas morphological profiling is an untargeted, systems biology approach.
Immunocytochemistry (ICC)
ICC relies on the specific binding of antibodies to target antigens, which are then visualized via chromogenic or fluorescent detection [45]. In neural cultures, this typically involves using antibodies against markers like β-III-tubulin (TUJ1) for neurons, GFAP for astrocytes, or SMI312 for axons and MAP2 for dendrites [48]. The output is semi-quantitative data on the presence, distribution, and localization of these specific targets.
High-Content Morphological Profiling
Morphological profiling automates the acquisition and analysis of microscopic images to extract vast numbers of quantitative features describing cell morphology, texture, and spatial relationships. Techniques like Cell Painting use a panel of fluorescent dyes to label multiple cellular compartments (e.g., nucleus, endoplasmic reticulum, Golgi apparatus, cytoskeleton, and mitochondria) [46] [49]. Subsequent image analysis using software like CellProfiler or convolutional neural networks (CNNs) can quantify over 1,000 morphological features, creating a "morphological fingerprint" for the cell population [46]. This fingerprint can distinguish cell types and states with high accuracy, even in dense, mixed neural cultures [49].
Table 1: Fundamental Characteristics of ICC and HCI-based Morphological Profiling
| Characteristic | Immunocytochemistry (ICC) | High-Content Morphological Profiling |
|---|---|---|
| Core Principle | Targeted antibody-antigen interaction | Untargeted, panoptic staining of cellular compartments |
| Primary Output | Semi-quantitative data on specific protein targets | Multiparametric quantitative data (1,000+ features) on cell morphology |
| Typical Markers | TUJ1, MAP2, SMI312, GFAP [48] | Multiplexed dyes for nuclei, cytoplasm, organelles [46] |
| Data Analysis | Manual scoring or basic intensity/area quantification | Automated feature extraction (CellProfiler) & machine learning (CNN) [48] [49] |
| Throughput | Low to medium | High to very high (automated) |
| Key Advantage | High molecular specificity | Unbiased, systems-level view; high content |
Performance Comparison in Neural Culture Validation
Direct and indirect comparisons from recent studies demonstrate the relative performance of these methodologies in key applications for neural culture validation.
Cell Type Identification and Classification Accuracy
A critical validation step is identifying and quantifying the different cell types within a mixed neural culture. A landmark study demonstrated that a Cell Painting approach combined with a Convolutional Neural Network (CNN) could recognize neural cell types in dense, mixed cultures with a fidelity exceeding 96% accuracy [49]. This "nucleocentric" profiling approach maintained high accuracy even in very dense cultures, where traditional cell segmentation and analysis often fail.
In contrast, while ICC can identify cell types, its classification is binary (positive/negative for a marker) and relies on the researcher pre-selecting the correct markers. It does not intrinsically provide a multivariate, quantitative profile of the cell's state.
Detecting Phenotypic Shifts in Disease Models
Morphological profiling excels at detecting subtle, complex phenotypic changes. In a model of Alzheimer's disease using SORL1-deficient human neural progenitor cells (NPCs), Cell Painting robustly distinguished the mutant NPCs from isogenic wild-type controls based on their morphological signatures [46]. Furthermore, the assay screened a 330-compound library and identified 16 compounds that reversed the mutant morphological signature, demonstrating its utility in phenotypic drug discovery.
ICC-based analysis in similar contexts typically focuses on a predefined set of markers, such as measuring endosome size, potentially missing broader, more subtle morphological alterations captured by Cell Painting [46].
Quantifying Neurite Outgrowth and Synapse Formation
Both techniques can be applied to study neuronal maturation and neurotoxicity, but with different depths of information. A dedicated HCI protocol for human iPSC-derived neurons uses automated microscopy to quantify the morphology of dendrites and axons in a high-throughput system [48]. It can extract specific parameters like neurite outgrowth, number of processes, and branches.
ICC for neurite outgrowth (e.g., using TUJ1 and MAP2) provides the foundational images for such analysis. However, traditional ICC analysis often lacks the high-throughput, multiparametric extraction capabilities of a dedicated HCI pipeline. HCI can also be extended to more complex 3D cultures, where confocal z-stacks are analyzed to quantify the total volume of neurites and number of branching points in three dimensions [50].
Table 2: Quantitative Performance Comparison from Key Studies
| Application | Morphological Profiling Performance | ICC Context & Limitations |
|---|---|---|
| Cell Type Classification | 96% accuracy using CNN on Cell Painting data [49] | Relies on pre-defined markers; no inherent classification algorithm. |
| Phenotypic Reversion Screening | 16 hits identified from 330-compound library in SORL1-/- NPCs [46] | Possible, but typically limited to a few pre-defined readouts (e.g., endosome size). |
| Neurite Outgrowth in 3D | Quantification of total neurite volume and branching points in hydrogel [50] | Provides images but 3D quantification is complex and less standardized. |
| Toxicity Screening (IC50) | Measured IC50 for neurite outgrowth (e.g., Methyl Mercury IC50 = 5 nM) [50] | Possible, but often with lower throughput and fewer concurrent parameters. |
Experimental Protocols for Key Applications
This protocol quantifies axonal and dendritic morphology within a high-throughput system.
- Cell Seeding and Culture: Coat a black-wall, clear flat-bottom 384-well plate with Geltrex (1:100 dilution) and incubate for 1 hour at 37°C. Seed dissociated iPSC-derived neurons (e.g., induced by NGN2 expression) in neuronal medium (Neurobasal medium supplemented with B-27, GlutaMAX, BDNF, NT-3, Laminin, and Doxycycline).
- Fixation and Staining: At the desired time point (e.g., 5 days post-seeding), fix cells with 4% PFA for 15 minutes. Permeabilize and block with a solution containing 3% BSA and 0.5% Triton X-100. Stain with primary antibodies: anti-SMI312 (axons, 1:500) and anti-MAP2 (dendrites, 1:1000), followed by appropriate fluorescent secondary antibodies (e.g., Alexa Fluor 568 for axons, CF 488A for dendrites). Counterstain nuclei with Hoechst 33342.
- Image Acquisition and Analysis: Acquire images using an HCA system (e.g., Operetta CLS). Use open-source software CellProfiler to create an analysis pipeline. The pipeline identifies nuclei, classifies neurons based on cytoskeletal staining, and performs neurite tracing to extract metrics for individual neurons, including total neurite length per neuron, number of branches, and number of processes.
This protocol generates a multivariate morphological profile for distinguishing NPC states.
- Cell Culture and Staining: Plate wild-type and mutant NPCs (e.g., SORL1-/-) in 96-well plates. Fix cells and perform the Cell Painting assay using a multiplexed dye set: Hoechst 33342 (nuclei), Phalloidin (cytoskeleton/filamentous actin), Concanavalin A (endoplasmic reticulum), WGA (Golgi apparatus), and MitoTracker (mitochondria). SYTO 14 can be used to stain RNA.
- Image Acquisition: Acquire high-resolution images in five or six fluorescent channels using a high-content imager.
- Image Analysis and Feature Extraction: Process images using CellProfiler software. Identify individual cells and extract morphological, intensity, and texture features for each compartment across all channels, resulting in a vector of >1,000 features per cell.
- Data Analysis and Machine Learning: Use dimensionality reduction (e.g., UMAP) and machine learning classifiers (e.g., Random Forest, CNN) to build a model that distinguishes cell types or states based on their morphological profiles. For drug screening, identify compounds that shift the mutant profile toward the wild-type state.
Diagram 1: Cell Painting Workflow for HCI
This protocol characterizes neuronal development in a more physiologically relevant 3D matrix.
- 3D Cell Culture: Use a synthetic PEG-based hydrogel (e.g., 3DProSeedTM) pre-casted in a 96-well plate. Seed human iPSC-derived neural cells (e.g., CNS.4U mix of neurons and astrocytes) on the hydrogel surface at 40,000 cells/well. Cells will penetrate the matrix and form 3D neurite networks over 14 days.
- Compound Treatment: For toxicity screening, add compounds 72 hours post-seeding. Refresh media with compounds every 2 days.
- Staining and Imaging: At endpoint, fix cells with 4% formaldehyde, permeabilize with 0.1% Triton X-100, and stain with antibody against β-III-tubulin (TUJ1, neuronal marker) and Hoechst (nuclei). Acquire confocal z-stacks (e.g., 11-33 planes, 5-10 μm apart) using a system like the ImageXpress Micro Confocal.
- 2D and 3D Analysis: Analyze 2D maximum projection images using a standard neurite outgrowth algorithm. For more accurate 3D quantification, use a 3D analysis module (e.g., in MetaXpress software) to connect objects across z-planes. Key readouts include total neurite volume, number of branches per well, and cell viability.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Reagent Solutions for High-Content Imaging in Neural Cultures
| Reagent / Material | Function / Application | Example Products / Markers |
|---|---|---|
| Cell Lines | Biologically relevant model systems | iPSC-derived neurons, NPCs, CNS.4U co-culture [48] [50] |
| Multiwell Plates | Vessel for high-throughput culture & imaging | Black-walled, μClear bottom 384-well plates [48] |
| Cytoskeletal Markers | Identify neurons and quantify neurites | β-III-tubulin (TUJ1), MAP2, SMI312 [48] |
| Cell Painting Dyes | Panoptic labeling of cellular compartments | Hoechst (DNA), Phalloidin (F-actin), Con A (ER), WGA (Golgi), MitoTracker [46] |
| Fixation & Permeabilization | Preserve cell structure and enable staining | 4% Paraformaldehyde (PFA), Triton X-100 [48] |
| HCA Imaging System | Automated image acquisition | Operetta CLS, ImageXpress Micro Confocal, CellInsight [48] [50] |
| Image Analysis Software | Quantitative feature extraction & analysis | CellProfiler, HCS Studio, MetaXpress [48] [46] |
| 3D Hydrogel Matrix | Physiologically relevant 3D cell culture | 3DProSeedTM PEG-based hydrogel [50] |
Integrated Analysis and Decision Framework
The choice between immunocytochemistry and high-content morphological profiling is not a simple binary decision but depends on the research question's stage and scope.
Diagram 2: Decision Framework for Method Selection
Opt for Immunocytochemistry (ICC) when: The research question is focused on validating the expression or localization of a specific, known protein (e.g., confirming neuronal identity with TUJ1). ICC is also a practical choice when resources for high-content instrumentation and advanced bioinformatics are limited.
Opt for High-Content Morphological Profiling when: The goal is an unbiased discovery of phenotypic differences, such as in disease modeling or functional genomics. It is the superior method for high-throughput compound screening where the mechanism of action may be unknown, and for detecting complex, multivariate phenotypes that cannot be captured by a few markers.
Use an Integrated Approach: The most powerful strategy often combines both. Morphological profiling can identify hit compounds or genetic perturbations in a primary screen. Follow-up analysis using targeted ICC on these hits can then provide mechanistic insight by validating specific molecular changes, creating a complementary and highly effective workflow [51].
Standardized ICC Protocols for Synaptic Connectivity Analysis (e.g., Pre/Post-Synaptic Marker Colocalization)
The validation of neural cultures, whether for basic research or the development of cell-based therapies, requires precise methods to confirm the presence and density of synaptic connections. Within this context, immunocytochemistry (ICC) has emerged as a foundational technique, allowing researchers to visualize and quantify synapses through the colocalization of specific pre- and postsynaptic markers. This guide provides an objective comparison of standardized ICC protocols for synaptic connectivity analysis, framing them as a counterpart to the emerging methodology of morphological profiling. While ICC offers targeted, protein-specific validation of synapses, morphological profiling uses high-content imaging and machine learning to classify cell types and states based on their physical characteristics, providing a broader, non-destructive assessment of culture composition [21] [1].
Core Principles of Synapse Quantification by ICC
In immunocytochemical terms, a synapse is defined by the immediate apposition of a presynaptic marker—typically a protein associated with synaptic vesicles—and a postsynaptic marker, often a scaffolding protein or receptor complex located in the postsynaptic density [52] [53]. The quantification of these colocalization events can be performed manually, in a semi-automated fashion, or through fully automated algorithms, which detect fluorescent puncta and define a synapse based on the overlap or proximity of the two signals within a defined distance threshold [52].
Table 1: Common Pre- and Postsynaptic Markers for ICC
| Synaptic Compartment | Marker Name | Primary Function | Nature of Protein |
|---|---|---|---|
| Presynaptic | Synapsin | Synaptic vesicle-associated phosphoprotein, regulates neurotransmitter release | Cytoplasmic [54] |
| Presynaptic | Synaptophysin | Major integral membrane protein of synaptic vesicles | Membrane [54] |
| Presynaptic | VGLUT1 | Loads glutamate into synaptic vesicles | Cytoplasmic [54] |
| Postsynaptic | PSD-95 | Main scaffolding protein in the postsynaptic density of excitatory synapses | Cytoplasmic [54] |
| Postsynaptic | Gephyrin | Primary scaffolding protein for inhibitory receptors | Cytoplasmic [54] |
| Postsynaptic | Homer1 | Scaffolding protein that links metabotropic glutamate receptors to other PSD proteins | Cytoplasmic [54] |
Comparative Analysis of Synaptic Quantification Methods
Different methodologies for synapse quantification offer varying degrees of throughput, objectivity, and technical demand. The table below compares three common approaches.
Table 2: Comparison of Synaptic Quantification Methodologies
| Method | Key Features | Advantages | Limitations | Suitability |
|---|---|---|---|---|
| Manual / Semi-Automated Counting | Relies on researcher identification or software-assisted puncta counting, often from 2D maximum intensity projections [52] [53]. | Accessible (e.g., using ImageJ plugins like Puncta Analyzer) [53]. | Work-intensive; prone to user bias and inter-lab variation; lower throughput [52]. | Lower-throughput studies; labs without specialized analysis software. |
| Fully Automated 3D Quantification | Algorithm-based detection of pre- and postsynaptic elements and their colocalization in three dimensions [52]. | High-throughput; unbiased; batch processing of large datasets; accounts for 3D structure [52]. | Requires custom software and parameter optimization; can be a "black box" [52]. | High-content screening; large-scale studies of synaptogenesis. |
| Morphological Profiling / Cell Painting | Uses high-content imaging and machine learning (e.g., CNNs) to classify cell types and states based on morphological fingerprints [1]. | Non-destructive; fast; can predict culture composition without specific markers [21] [1]. | Does not directly quantify synapses; requires initial training with validated datasets [1]. | Initial quality control of neural cultures; non-destructive validation. |
Detailed Standardized ICC Protocol for Dissociated Neuronal Cultures
The following protocol, adapted from established methodologies, is designed for quantifying synapses in dissociated neuronal cultures grown on glass coverslips [52] [53].
Solutions and Reagents
- Antibody Buffer: 150 mM NaCl, 50 mM Tris-Base, 1% BSA, 100 mM L-lysine, 0.04% Azide; adjust pH to 7.4 and filter [53].
- Fixative: 4% paraformaldehyde (PFA) in PBS, pre-warmed to 37°C [53].
- Blocking Buffer: 50% Normal Goat Serum, 50% Antibody Buffer, 0.2% Triton X-100 [53].
- Primary and Secondary Antibodies: Centrifuge before use to remove precipitates [53].
Step-by-Step Procedure
- Fixation: Remove culture media and add 4% PFA to cover cells. Incubate for 7 minutes at room temperature. Rinse 3x with PBS. Note: Cells should never be allowed to dry out. [53]
- Blocking: Apply blocking buffer for 30 minutes at room temperature to reduce non-specific binding. Rinse 3x with PBS [53].
- Primary Antibody Incubation: Prepare primary antibodies (e.g., mouse anti-Homer1 for postsynaptic, rabbit anti-Synapsin for presynaptic) in a solution of 90% antibody buffer and 10% NGS. Apply to coverslips and incubate overnight at 4°C in a humidified chamber. Rinse 3x with PBS [53].
- Secondary Antibody Incubation: Prepare fluorophore-conjugated secondary antibodies (e.g., Alexa Fluor 488 and 594) diluted 1:1000 in antibody buffer with 10% NGS. Apply for 2 hours at room temperature in the dark. Rinse 3-4x with PBS [53].
- Mounting: Mount coverslips onto glass slides using an anti-fade mounting medium containing DAPI. Seal edges with clear nail polish [53].
- Imaging: Image using a fluorescence microscope with a 63x or higher oil-immersion objective. For unbiased analysis, select fields systematically (e.g., via DAPI channel) and acquire high-resolution z-stacks [52] [53].
- Image Analysis: Use automated software to detect fluorescent puncta in each channel and identify colocalizations. Synapses are typically defined as pre- and postsynaptic signals falling within a distance threshold of 0.7 - 2.5 μm [52].
Successful synaptic connectivity analysis relies on a suite of well-validated reagents and computational tools.
Table 3: Key Research Reagent Solutions for Synaptic ICC
| Category | Item / Tool | Specific Function / Example |
|---|---|---|
| Validated Antibodies | Presynaptic Marker Kit | Contains multiple validated antibodies (e.g., against Synapsin, SNAP-25, VGLUT) for comprehensive presynaptic labeling [54]. |
| Postsynaptic Marker Kit | Contains multiple validated antibodies (e.g., against PSD-95, Gephyrin, Shank) for comprehensive postsynaptic labeling [54]. | |
| Cell Culture & Staining | Primary Neuronal Culture Systems | Dissociated hippocampal neurons are a well-characterized model for synaptogenesis studies [52]. |
| Cell Painting Dyes | A panel of fluorescent dyes (e.g., for nuclei, cytoskeleton, organelles) used in morphological profiling to generate a morphological fingerprint [1]. | |
| Image Analysis Software | Puncta Analyzer (ImageJ Plugin) | Semi-automated tool for colocalization studies in 2D datasets [53]. |
| Custom Automated 3D Analysis | Fully automated algorithms for 3D synapse detection and quantification, often with batch processing capabilities [52]. | |
| Convolutional Neural Networks (CNNs) | Deep learning models for high-accuracy cell type classification in dense cultures based on morphological features [1]. |
Integrated Workflow: Combining ICC and Morphological Profiling for Culture Validation
A robust strategy for neural culture validation can leverage the strengths of both ICC and morphological profiling. The latter serves as an initial, non-destructive quality control step, identifying and selecting organoids or culture regions with desired characteristics based on morphology alone, which strongly correlates with cellular composition [21]. Subsequently, targeted ICC analysis provides definitive, protein-specific confirmation of synaptic density and maturity.
This integrated approach enhances experimental reproducibility and is crucial for ensuring the safety and efficacy of neural cultures intended for therapeutic applications [21].
The pursuit of effective treatments for Parkinson's (PD) and Alzheimer's disease (AD) increasingly relies on patient-derived neuronal cultures, which provide physiologically relevant human models for studying disease mechanisms and screening potential therapeutics [55]. A significant challenge in this field, however, lies in the accurate and consistent validation of these complex cellular models. Variations in differentiation protocols, genetic background, and cellular heterogeneity can lead to inconsistent and potentially misleading results, hindering experimental reproducibility and translational success [1]. This has created an urgent need for robust quality control methods to characterize culture composition, purity, and maturity.
Two principal technological approaches have emerged to address this validation challenge: traditional immunocytochemistry (ICC) and the increasingly prominent morphological profiling. ICC identifies specific cell types through targeted molecular recognition of intracellular and surface proteins, providing high specificity but requiring predefined markers and being inherently destructive [10] [1]. In contrast, morphological profiling utilizes high-content imaging and computational analysis to classify cells based on their physical characteristics—size, shape, texture, and spatial organization—offering a potentially non-destructive, label-free, and unbiased assessment of cellular state and heterogeneity [10] [1] [56]. This guide provides a detailed comparison of these competing methodologies, evaluating their performance, applications, and suitability for preclinical research in Parkinson's and Alzheimer's disease.
Technological Face-Off: Morphological Profiling vs. Immunocytochemistry
Core Principles and Methodologies
Immunocytochemistry (ICC) operates on the principle of antibody-antigen recognition. Cells are fixed, permeabilized, and incubated with fluorescently-labeled antibodies targeting specific proteins (e.g., β-III-tubulin for neurons, GFAP for astrocytes). The presence and intensity of fluorescence serve as a direct readout for protein expression, allowing for precise identification of cell types and states [10]. For instance, the co-expression of specific surface markers like CD105, CD90, and CD73 is a established criterion for defining multipotent mesenchymal stromal cells, with the loss of CD73 correlating with early differentiation into flattened cells [10]. A typical ICC protocol involves seeding cells on glass slides, fixation with paraformaldehyde, blocking with serum, incubation with primary and secondary antibodies, and final imaging using fluorescence microscopy [10].
Morphological Profiling, particularly the Cell Painting (CP) assay, takes a holistic, unsupervised approach. It uses a panel of fluorescent dyes to non-specifically stain multiple cellular compartments: the nucleus (e.g., Hoechst), nucleoli (e.g., with a RNA-binding dye), endoplasmic reticulum (e.g., Concanavalin A), cytoskeleton (e.g., phalloidin), and Golgi apparatus [1]. High-content imaging captures the morphological landscape, and subsequent computational feature extraction—quantifying shape, intensity, texture, and inter-object relationships—generates a high-dimensional "morphological fingerprint" for each cell. Machine learning classifiers, such as convolutional neural networks (CNNs), are then trained to identify cell types or states based on these fingerprints alone, without prior molecular knowledge [1].
Table 1: Core Methodological Comparison
| Feature | Immunocytochemistry (ICC) | Morphological Profiling |
|---|---|---|
| Primary Readout | Specific protein expression | Whole-cell morphological features |
| Throughput | Lower (multi-step, destructive) | Higher (amenable to automation) |
| Bias | High (requires pre-selected markers) | Low (unbiased, discovery-oriented) |
| Cost per Assay | Moderate to High (antibody costs) | Low (inexpensive dyes) |
| Cell Fate Post-Assay | Fixed/dead (destructive) | Potentially live (if label-free) |
| Key Strength | High molecular specificity | Captures cellular heterogeneity |
Performance and Accuracy Metrics
Recent studies have directly benchmarked the classification performance of morphological profiling against established ICC-based identification. In a landmark study using neural cell lines (1321N1 astrocytoma and SH-SY5Y neuroblastoma), a morphological profiling pipeline achieved a remarkable 96% accuracy in distinguishing cell types in mixed cultures [1]. This performance significantly outperformed a traditional machine learning approach (Random Forest) using hand-crafted morphological features, which only reached an F-score of 0.75 [1]. This demonstrates the power of deep learning in extracting relevant biological signals from complex image data.
The application to more physiologically relevant, patient-derived models further underscores its utility. When applied to iPSC-derived neural cultures to distinguish postmitotic neurons from neural progenitors, the cell-based morphological prediction significantly outperformed a simple classification based on the population-level time in culture (96% vs. 86% accuracy) [1]. Furthermore, the method could unequivocally discriminate neurons from microglia, and even distinguish activated from non-activated microglial states, albeit with lower accuracy for the latter, more subtle distinction [1].
Table 2: Experimental Performance Data from Key Studies
| Experimental Context | Cell Types/States Distinguished | Reported Accuracy/Metric | Key Finding |
|---|---|---|---|
| Neural Cell Lines [1] | Astrocytoma (1321N1) vs. Neuroblastoma (SH-SY5Y) | >96% (CNN-based classification) | Superior to feature-based Random Forest (F-score: 0.75) |
| iPSC-Derived Neural Cultures [1] | Postmitotic Neurons vs. Neural Progenitors | 96% (cell-based prediction) | Outperformed time-in-culture prediction (86%) |
| iPSC-Derived Neural Cultures [1] | Neurons vs. Microglia | Unequivocal discrimination | Robust to microglial reactivity state |
| hMSC Subpopulations [10] | Rapidly self-renewing (RS) vs. Flattened (FC) cells | Correlation with CD105/CD90/CD73 loss | FC cells mainly lack CD73, linking morphology to marker expression |
Application in Alzheimer's and Parkinson's Disease Modeling
Both techniques are instrumental in modeling the pathological hallmarks of AD and PD. ICC remains the gold standard for identifying and quantifying key protein aggregates: amyloid-β (Aβ) plaques and hyperphosphorylated Tau in neurofibrillary tangles in AD [57] [58], and alpha-synuclein (αSyn) in Lewy bodies in PD [59] [60]. These pathologies are defining features in post-mortem brain tissue and are recapitulated in advanced in vitro models like brain organoids [55].
Morphological profiling offers a complementary, systems-level view. It can detect early, pre-pathological changes in cellular state driven by genetic risk factors. For example, in a sophisticated "miBrain" model that incorporates all six major brain cell types, researchers demonstrated that APOE4 astrocytes (a major genetic risk factor for AD) only exhibited immune reactivity when cultured in the multicellular miBrain environment, not in isolation. Furthermore, cross-talk between APOE4 astrocytes and microglia was found to be required for the accumulation of phosphorylated tau pathology [5]. Such complex, multi-cellular interactions are difficult to dissect with ICC alone but are readily captured by morphological and functional analyses in integrated models.
Furthermore, the common neuroinflammatory pathways in AD and PD—involving the activation of microglia and astrocytes and the release of pro-inflammatory cytokines—lead to characteristic and measurable morphological shifts, such as astrocyte hypertrophy, which can be quantified via profiling [60].
The Scientist's Toolkit: Essential Research Reagent Solutions
The successful implementation of these validation strategies depends on a suite of essential research reagents and tools.
Table 3: Key Research Reagent Solutions
| Item Category | Specific Examples | Function in Validation |
|---|---|---|
| Cell Culture & Differentiation | MEM Alpha GlutaMAX, B27 Supplement, Matrigel | Supports growth and neural differentiation of iPSCs [10] [55]. |
| Immunocytochemistry Reagents | Antibodies: CD105, CD90, CD73; β-III-tubulin; GFAP; Iba1; Aβ; p-Tau; αSyn | Molecular identification of cell types, stemness, and disease pathologies [10] [1] [58]. |
| Morphological Profiling Dyes | Hoechst (DNA), Phalloidin (F-actin), Concanavalin A (ER), RNA-binding dyes (Nucleoli) | Stains multiple organelles to create a whole-cell morphological profile for Cell Painting [1]. |
| Image Analysis Software | MetaMorph, ImageJ/FIJI, Custom CNN scripts (e.g., ResNet) | Automates cell segmentation, feature extraction, and classification [1] [56]. |
| Advanced Disease Models | Brain Organoids, "miBrains" (Multicellular Integrated Brains) | Provides a physiologically complex 3D environment for disease modeling and therapy testing [5] [61] [55]. |
Experimental Design: A Tiered Workflow for Culture Validation
A robust strategy for validating patient-derived neuronal cultures often involves a tiered workflow that leverages the strengths of both morphological profiling and ICC.
Step 1: Rapid, Non-Destructive Quality Control. Morphological profiling serves as an initial, rapid screen. High-content imaging of live or minimally stained cultures can quickly assess confluency, overall culture health, and major contamination (e.g., fibroblast overgrowth). Automated analysis of neurite outgrowth and branching—a strong indicator of neuronal maturity and health—can be efficiently performed using software like MetaMorph [56].
Step 2: In-depth Characterization of Composition and State. For a more detailed analysis, the full Cell Painting assay is performed. The extracted morphological fingerprints are used to classify cells into types (neurons, astrocytes, microglia) and potentially identify aberrant states. This step is crucial for quantifying heterogeneity within a culture.
Step 3: Targeted, High-Specificity Validation. Based on the results of morphological profiling, targeted ICC is deployed to confirm specific identities. For example, if a subpopulation of cells is morphologically suspect for being reactive astrocytes, ICC for GFAP and C3 (an A1 astrocyte marker) would provide definitive confirmation [60]. Similarly, the presence of disease-relevant proteins like p-Tau or αSyn is definitively confirmed via ICC.
This tiered approach balances speed and comprehensiveness, using morphological profiling as a wide-angle lens to survey the entire landscape and ICC as a telephoto lens for high-magnification detail on critical features.
The validation of patient-derived neuronal models for Alzheimer's and Parkinson's research is a critical step that directly impacts the translational value of preclinical findings. While immunocytochemistry remains the undisputed method for definitive identification of specific molecular targets and well-characterized pathologies, morphological profiling has emerged as a powerful, high-throughput partner that excels in capturing cellular heterogeneity, identifying novel states, and detecting subtle, system-wide changes in response to genetic risks or chemical perturbations.
The future of neural culture validation lies not in choosing one method over the other, but in strategically integrating them. A tiered workflow that uses morphological profiling for initial screening and hypothesis generation, followed by targeted ICC for confirmation and deep molecular insight, provides a comprehensive, efficient, and robust framework. This synergistic approach will be essential for accelerating the development of reliable, human-relevant models and, ultimately, effective therapies for neurodegenerative diseases.
Cerebral organoids have emerged as groundbreaking tools in neuroscience, providing complex in vitro models that mimic aspects of human brain development and function. These three-dimensional structures, derived from pluripotent stem cells, self-organize into architectures that recapitulate certain regions of the human brain, offering unprecedented opportunities for studying neurodevelopmental processes, neurological diseases, and neurotoxicity [62] [63]. Unlike traditional 2D cultures or simpler 3D models, cerebral organoids recreate a physiologically relevant cellular microenvironment that enhances cell-cell and cell-matrix interactions, fostering improved differentiation and maturation [62]. This biological relevance makes them particularly valuable for disease modeling, drug screening, and toxicology studies in both academic and industrial settings.
However, the significant potential of cerebral organoids is hampered by substantial challenges related to quality and reproducibility. Discrepancies in morphology, size, cellular composition, and cytoarchitectural organization limit their reliability and broader application [64] [62]. This variability stems from the stochastic nature of stem cell differentiation and spontaneous self-organization within organoids, resulting in inconsistencies that compromise reproducible scientific results [62]. Within a single batch, some organoids display optimal morphology with dense structure and well-defined borders, while others may be poorly compacted, degrade over time, develop suboptimal cystic cavities, or contain necrotic cores [62]. These challenges are exacerbated by a lack of standardized criteria for organoid generation, culture, and characterization across research laboratories.
This article explores a newly developed Quality Control (QC) framework specifically designed for 60-day cortical organoids, which represents a critical intermediate stage of maturation characterized by the presence of neural progenitors, neurons, and astrocytes [62] [63]. We will examine how this scoring system addresses reproducibility challenges through a hierarchical evaluation approach, compare its methodology with traditional validation techniques like immunocytochemistry, and provide practical implementation guidance for researchers seeking to adopt standardized quality assessment protocols for their 3D neural culture systems.
A Novel QC Framework: Hierarchical Scoring for 60-Day Cortical Organoids
Framework Architecture and Core Components
To address the pressing need for standardization in cerebral organoid research, a comprehensive QC framework has been developed specifically for 60-day cortical organoids [64] [62]. This system employs a structured scoring methodology that evaluates organoids across five critical criteria, each subdivided into specific indices with scores ranging from 0 (low quality) to 5 (high quality) [62]. The framework is designed hierarchically, prioritizing non-invasive assessments initially, with more in-depth analyses reserved for organoids that meet preliminary thresholds [64].
The five primary criteria include:
- Morphology: Assessing compactness, border integrity, and absence of cysts [63]
- Size and Growth Profile: Ensuring proper developmental dynamics [62]
- Cellular Composition: Verifying presence of expected neural populations [62]
- Cytoarchitectural Organization: Evaluating structural integrity and organization of neural layers [62] [63]
- Cytotoxicity: Measuring DNA damage and cell viability [62]
This hierarchical organization allows for efficient resource allocation by quickly excluding low-quality organoids from further analysis while focusing detailed characterization efforts on specimens that meet baseline quality standards [62].
Implementation Workflow: Initial and Final QC Assessment
The QC framework operates through two distinct applications designed for different stages of research workflows. The Initial QC relies exclusively on the first two non-invasive criteria (morphology and size) to determine organoid eligibility before beginning a study [62]. This pre-study evaluation enables researchers to select only high-quality starting materials for their experiments, reducing confounding variables introduced by specimen variability.
The Final QC incorporates all five scoring criteria for comprehensive analysis at a study's conclusion [62]. This post-study assessment provides a complete quality profile of the organoids used in experimentation, allowing researchers to contextualize their findings against quality metrics and improving cross-study comparability.
To validate this scoring methodology, researchers exposed 60-day cortical organoids to increasing doses of hydrogen peroxide to generate specimens with varying quality levels [62]. The QC system successfully discriminated between these quality levels, demonstrating its robustness and sensitivity in detecting quality variations induced by oxidative stress [64] [62].
Figure 1: Hierarchical QC Workflow for Cerebral Organoids. The framework employs a two-stage assessment process beginning with non-invasive criteria (Initial QC) before proceeding to comprehensive analysis (Final QC) for specimens meeting quality thresholds [62].
Comparative Analysis: Morphological Profiling vs. Immunocytochemistry
Methodological Comparison for Neural Culture Validation
The validation of neural cultures traditionally relies heavily on immunocytochemistry (ICC) approaches, which provide detailed molecular information about cellular composition and protein expression. While valuable, these methods have limitations for routine quality assessment, particularly in 3D systems. The novel QC framework offers an alternative approach centered on morphological profiling, creating complementary but distinct methodologies for organoid validation.
Table 1: Methodological Comparison: Morphological Profiling vs. Immunocytochemistry
| Parameter | Morphological Profiling QC Framework | Traditional Immunocytochemistry |
|---|---|---|
| Primary Focus | Macroscopic structure and growth patterns | Molecular and protein-level characterization |
| Key Metrics | Morphology, size, growth profile, structural organization | Protein expression, cellular composition, marker localization |
| Throughput | High (especially initial QC steps) | Moderate to low |
| Destructiveness | Partially non-destructive | Typically destructive |
| Cost Efficiency | Higher for initial screening | Lower due to reagent costs |
| Temporal Resolution | Can monitor development over time | Single time point assessment |
| Standardization Potential | High (quantitative scoring system) | Moderate (subject to protocol variations) |
| Information Depth | Structural and organizational integrity | Molecular composition and cell type identification |
| Technical Expertise Required | Moderate | High for standardization |
| Scalability | Highly scalable for industrial applications | Less scalable for high-throughput screening |
Integration Potential and Complementary Applications
Rather than representing competing methodologies, morphological profiling and immunocytochemistry offer complementary strengths that can be strategically integrated within research workflows. The QC framework serves as an efficient screening tool that can identify representative specimens for more detailed molecular characterization via ICC [62]. This integrated approach maximizes resource efficiency by reserving labor-intensive and costly immunocytochemical analyses for organoids that have already demonstrated structural competence.
The hierarchical nature of the QC framework specifically facilitates this integrated validation approach. As researchers proceed through the scoring system, they naturally transition from macroscopic morphological assessments to more detailed analyses of cellular composition and cytoarchitectural organization, which can be complemented and verified through targeted immunocytochemistry [62]. This methodological synergy addresses the limitation of traditional ICC approaches, which often lack standardization and face challenges in transposition from 2D to 3D culture systems [62].
Experimental Validation: Protocol and Application
Experimental Design for QC Framework Validation
The robustness of the QC scoring system was validated through controlled experiments exposing 60-day cortical organoids to graded doses of hydrogen peroxide to induce a range of quality outcomes [62]. This experimental approach generated specimens with quantifiable variations in quality parameters, allowing rigorous testing of the framework's ability to discriminate between different quality levels.
Table 2: Key Reagent Solutions for Cerebral Organoid QC Assessment
| Reagent/Category | Primary Function | Specific Application in QC |
|---|---|---|
| Hydrogen Peroxide (H₂O₂) | Inducer of oxidative stress | Generation of quality variants for validation [62] |
| Differentiation Media | Support neural differentiation | 60-day cortical organoid maturation [62] [63] |
| Viability Assays | Cell health assessment | Cytotoxicity scoring [62] |
| Molecular Markers | Cell type identification | Cellular composition verification [62] |
| Imaging Contrast Agents | Structural visualization | Morphology and organization assessment [62] |
| Fixation Reagents | Sample preservation | Preparation for detailed analysis [62] |
The experimental protocol followed this structured approach:
- Organoid Selection: 60-day cortical organoids were first selected using the Initial QC method based on non-invasive morphology and size criteria [62]
- Treatment Application: Selected organoids were exposed to graduated concentrations of H₂O₂ to induce varying degrees of quality compromise [62]
- Recovery Period: Following exposure, organoids underwent a one-week recovery period to allow quality variations to manifest [62]
- QC Assessment: Both exposed and non-exposed control organoids underwent comprehensive evaluation using the complete Final QC scoring system [62]
- System Validation: The framework's performance was quantified by its ability to accurately discriminate organoid qualities across the induced quality spectrum [62]
This validation protocol demonstrated the QC system's robustness by accurately classifying organoids according to their quality status, confirming its utility for standardized assessment [64] [62].
Figure 2: Experimental Workflow for QC Framework Validation. The validation protocol involves initial quality screening, controlled quality challenge through H₂O₂ exposure, recovery period, and comprehensive final assessment to verify classification accuracy [62].
Quantitative Scoring Metrics and Thresholds
The QC framework employs a precise scoring system with defined thresholds for each quality criterion. For each index, organoids receive scores from 0-5, with minimum thresholds established for progression through the hierarchical assessment [62]. Organoids failing to meet minimum scores for any criterion are categorized as low-quality and excluded from further evaluation, while those meeting composite thresholds across multiple indices receive appropriate quality classifications [62].
The specific scoring metrics include:
- Morphological Index: Quantitative assessment of compactness, border integrity, and cystic structure formation
- Size and Growth Metrics: Comparison against expected developmental trajectories for 60-day cortical organoids
- Cellular Composition Scores: Evaluation of neural population ratios and marker expression patterns
- Cytoarchitectural Organization: Scoring of structural integrity and layer formation
- Cytotoxicity Measurements: Quantification of cell viability and DNA damage markers
This quantitative approach minimizes observer bias and enables objective, reproducible quality assessments that enhance consistency and comparability across research groups and studies [62].
Implications for Research and Industry
The development of a standardized QC framework for cerebral organoids addresses critical barriers to broader adoption in both academic and industrial settings. By providing a common language and standardized metrics for quality assessment, this system enhances the reliability and interpretability of research findings, particularly in disease modeling, neurotoxicity testing, and drug screening applications [62] [65].
For pharmaceutical and toxicology applications, the scalability of this QC framework enables its implementation in industrial contexts where high-throughput screening and standardized protocols are essential [62] [63]. The hierarchical design supports efficient resource allocation by quickly identifying suboptimal specimens early in the testing pipeline, reducing costs associated with running comprehensive assays on low-quality materials [62]. This efficiency gain is particularly valuable in preclinical screening phases where large compound libraries are evaluated.
The framework's flexibility also allows for adaptation to different cerebral organoid types and developmental timepoints beyond the specific 60-day cortical organoids for which it was initially designed [62] [63]. This adaptability ensures broader utility across diverse research applications and experimental designs, supporting continued innovation in 3D neural culture systems while maintaining quality standards.
The quality control framework for cerebral cortical organoids represents a significant advancement in standardizing complex 3D neural culture systems. By integrating quantitative morphological profiling with hierarchical assessment protocols, this approach addresses critical reproducibility challenges while maintaining practical implementation feasibility. The comparative analysis with traditional immunocytochemistry methods highlights complementary strengths that can be strategically leveraged through integrated validation workflows.
As the field of 3D neural modeling continues to evolve, standardized quality assessment protocols will play an increasingly vital role in ensuring research reliability and translational applicability. The QC scoring system for cerebral organoids provides a robust foundation for these efforts, offering both academic and industrial researchers a validated tool for enhancing consistency, comparability, and confidence in their experimental models.
Overcoming Technical Hurdles: Optimization and Best Practices for Reliable Results
Immunocytochemistry (ICC) serves as a fundamental tool for visualizing protein localization and expression within cultured cells, providing critical insights into cellular function and heterogeneity. However, researchers consistently face challenges with artifacts and variability stemming from fixation methods, antibody specificity, and quantification approaches. These technical limitations become particularly problematic in advanced applications such as validating neural culture composition, where accurate cell-type identification is paramount. In recent years, morphological profiling based on high-content imaging has emerged as a complementary approach that may address some limitations of traditional antibody-based methods. This comparison guide objectively evaluates the performance of standard ICC protocols against emerging morphological profiling techniques, providing experimental data and methodologies to help researchers select the most appropriate validation strategy for their specific research context, particularly in neural culture applications.
Technical Comparison: ICC vs. Morphological Profiling
Table 1: Core Characteristics of ICC and Morphological Profiling Techniques
| Parameter | Immunocytochemistry (ICC) | Morphological Profiling |
|---|---|---|
| Basis of Identification | Specific antibody-antigen binding | Whole-cell morphotextural fingerprint |
| Multiplexing Capacity | Typically 2-4 targets simultaneously [12] | Unlimited in principle; limited by dye spectra [1] |
| Throughput | Medium (handling and incubation steps) | High (automated imaging and analysis) [1] |
| Quantification Approach | Semi-quantitative (intensity measurements) | Quantitative (multiparametric analysis) [1] |
| Primary Sources of Variability | Fixation, antibody specificity, permeabilization [66] [67] | Segmentation errors, dye variability |
| Cell Type Discrimination Accuracy | Dependent on antibody quality and specificity | >96% in controlled conditions [1] |
| Required Validation | Extensive antibody controls [68] [69] | Reference dataset validation |
Table 2: Performance Comparison in Neural Culture Applications
| Application Context | ICC Performance | Morphological Profiling Performance |
|---|---|---|
| Dense Mixed Cultures | Challenging due to antibody penetration issues [66] | High accuracy (>96%) maintained [1] |
| Neural Progenitor Identification | Good with validated markers (e.g., Nestin, SOX2) | 96% accuracy vs. 86% with culture time alone [1] |
| Subcellular Localization | Excellent with optimized protocols [70] [71] | Limited to cellular and subcellular regions |
| Cell State Differentiation | Possible with multiple markers | Distinguishes activated vs. non-activated states [1] |
| Primary Neural Cells | Established protocols available [70] | Requires reference dataset development |
Fixation-Induced Artifacts
Fixation represents the initial critical step where significant artifacts can be introduced into ICC experiments. Paraformaldehyde (PFA) fixation preserves cell morphology through protein cross-linking but can mask epitopes, while organic solvents like methanol simultaneously fix and permeabilize cells but may disrupt cellular structures [71]. The optimal fixation protocol must be determined empirically for each antigen-cell type combination, as longer incubation times can lead to over-fixed epitopes while shorter times may result in poor epitope preservation [71]. For neural cultures, a specialized 4% PFA fixative containing additives like MgCl₂ and EGTA helps preserve cytoskeletal structure and overall morphology, which is particularly important for maintaining intricate neural processes [70].
Antibody Specificity Challenges
Antibody specificity remains a fundamental concern in ICC, with a significant percentage of commercial antibodies lacking proper validation. One survey found that only 25-62% of antibodies used in scientific publications had adequate identification and specificity controls [67]. These "magic antibodies" – those with unverified specificity – represent a major source of irreproducibility in ICC experiments [67]. For neural cultures, where markers like β-III-tubulin, GFAP, and nestin are commonly used, improper antibody validation can lead to misidentification of cell types and erroneous conclusions about neural differentiation efficiency.
Table 3: Antibody Validation Methods and Their Applications
| Validation Method | Procedure | Evidence Level | Limitations |
|---|---|---|---|
| Knockout/Knockdown Models | Compare staining in WT vs. KO/KD cells [68] | Gold standard | Not always available; tissue-specific |
| Pre-adsorption Control | Pre-incubate antibody with immunogen [68] [69] | High for peptide antibodies | Less effective for whole protein immunogens |
| Isotype Control | Use non-immune immunoglobulin [69] | Identifies non-specific Fc binding | Does not confirm target specificity |
| Genetic Overexpression | Transfert cells to overexpress target [68] | Confirms recognition of intended target | Non-physiological expression levels |
| Multi-antibody Validation | Use multiple antibodies against different epitopes [68] | High with concordant results | Epitopes may be similarly inaccessible |
Quantification Variability
Quantification in ICC faces multiple challenges, including background fluorescence, non-specific binding, and cell-to-cell variability. For neural cultures, where cells exhibit complex morphologies with extensive processes, consistent segmentation and measurement present additional hurdles. The semi-quantitative nature of ICC intensity measurements means that results are often best interpreted as relative comparisons rather than absolute quantifications [12]. Morphological profiling approaches address some of these limitations through multiparametric analysis of shape, intensity, and texture features, enabling more robust quantitative classification of cell types in mixed neural cultures [1].
Experimental Protocols for Method Comparison
Standard ICC Protocol for Neural Cells
The following protocol has been optimized for neural cells and includes critical steps to minimize artifacts:
Day 1: Cell Preparation and Plating
- Use German glass coverslips cleaned with 1% Liquinox and 1M HCl for optimal cell adhesion [70].
- Coat coverslips with poly-D-lysine (10 µg/ml) for 5 minutes followed by laminin (20 µg/ml) in EMEM for 4 hours or overnight at 37°C [70].
- Seed human neural stem/precursor cells at 100,000-300,000 cells/ml and allow to adhere for at least 4 hours [70].
Day 2: Fixation, Permeabilization, and Primary Antibody Incubation
- Fix cells with pre-warmed 4% PFA fixative (with MgCl₂ and EGTA) for 10 minutes at room temperature [70].
- Wash 3 times with PBS, 5 minutes each wash.
- Permeabilize with 0.3% Triton X-100 in PBS for 5 minutes for intracellular targets [70].
- Wash 3 times with PBS, 5 minutes each.
- Block with 5% BSA in PBS for 1 hour at room temperature to reduce non-specific binding [70].
- Incubate with primary antibody diluted in 1% BSA/PBS overnight at 4°C using inverted coverslip method (25 µl per 25mm coverslip) [70].
Day 3: Secondary Antibody and Visualization
- Wash 3 times with PBS, 5 minutes each.
- Incubate with species-appropriate secondary antibody conjugated to fluorophore in 1% BSA for 2 hours in the dark [70].
- Wash 2 times with PBS, 5 minutes each.
- Incubate with Hoechst nuclear stain (2 µg/ml) for 1 minute [70].
- Wash once with PBS for 5 minutes.
- Mount coverslips with anti-fade mounting medium such as Vectashield [70].
Morphological Profiling Protocol for Neural Culture Validation
This protocol adapts the cell painting approach for neural cell identification:
Cell Staining
- Stain cells with a 4-channel fluorescent dye set: Hoechst 33342 (nuclei), Concanavalin A (glycoproteins), Wheat Germ Agglutinin (membranes), and Phalloidin (actin cytoskeleton) [1].
- Fix cells with 4% PFA as in standard ICC protocol.
Image Acquisition
- Acquire images using high-content confocal microscopy with consistent exposure across samples [1].
- Collect multiple fields per well to ensure statistical power.
Image Analysis and Classification
- Segment cells using convolutional neural networks to identify individual cells, even in dense cultures [1].
- Extract morphotextural features describing shape, intensity, and texture for nucleus, cytoplasm, and whole cell [1].
- Train ResNet convolutional neural network classifier on reference datasets of pure cell types [1].
- Apply classifier to mixed cultures for cell type identification.
- Validate classification accuracy against immunostaining for key markers [1].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Key Reagent Solutions for ICC and Morphological Profiling
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Poly-D-Lysine | Enhances cell adhesion to coverslips | Essential for primary neural cells [70] |
| Laminin | Improves neural cell attachment and differentiation | Used after poly-D-lysine coating [70] |
| Paraformaldehyde (4%) | Protein cross-linking fixative | Preserves morphology; may mask epitopes [70] [71] |
| Triton X-100 | Detergent for membrane permeabilization | Optimize concentration (0.1-0.3%) to balance access vs. preservation [71] |
| Bovine Serum Albumin (BSA) | Blocking agent reduces non-specific binding | Use at 2-10% in PBS; compatible with most antibodies [71] |
| Normal Serum | Alternative blocking agent | Use serum from secondary antibody host species [71] |
| Hoechst 33342 | Nuclear counterstain | Enables cell identification and segmentation [70] [1] |
| Fluorophore-conjugated Secondary Antibodies | Signal generation | Select based on microscope filter sets; consider brightness and photostability [12] |
| Anti-fade Mounting Medium | Preserves fluorescence | Essential for quantitative work; prevents photobleaching [70] |
| Cell Painting Dye Set | Multiplex morphological staining | Enables morphological profiling without antibodies [1] |
The choice between traditional ICC and emerging morphological profiling approaches for neural culture validation depends on specific research requirements. ICC provides superior subcellular resolution and direct protein identification when properly validated and optimized, but faces challenges with antibody specificity, artifacts, and limited multiplexing capacity. Morphological profiling offers high-throughput, multiparametric quantitative analysis that maintains accuracy even in dense cultures, making it particularly valuable for quality control of complex neural cultures. For robust neural culture validation, a combined approach leveraging the specificity of well-validated ICC protocols with the quantitative power of morphological profiling may provide the most comprehensive solution. Researchers should implement rigorous validation controls for either method and select the approach that best addresses their specific needs for resolution, throughput, and quantification.
Optimizing Segmentation and Feature Extraction in Dense, Mixed Neural Cultures
The adoption of human induced pluripotent stem cell (iPSC)-derived neural cultures has revolutionized neuroscience by providing physiologically relevant human models for studying brain development, dysfunction, and therapeutic interventions [1]. However, these cultures typically contain a complex mixture of cell types—including neurons, neural progenitors, astrocytes, and microglia—at high densities, creating significant challenges for accurate analysis [1] [72]. Traditional validation methods like immunocytochemistry and flow cytometry are often low-throughput, costly, and destructive, creating an urgent need for more efficient analytical pipelines [1].
This guide provides a comprehensive comparison of modern computational methods for segmenting and extracting morphological features from dense, mixed neural cultures. We objectively evaluate competing approaches—including traditional feature extraction, convolutional neural networks (CNNs), and novel algorithms—focusing on their performance characteristics, technical requirements, and suitability for different research scenarios. By framing this comparison within the broader thesis of morphological profiling versus immunocytochemistry for culture validation, we aim to equip researchers with the data needed to select optimal methodologies for their specific applications.
Comparative Analysis of Segmentation Methodologies
Accurate segmentation of individual cells in dense neural networks represents the foundational step in morphological analysis. We evaluated multiple computational approaches using standardized performance metrics.
Performance Comparison of Segmentation Algorithms
Table 1: Quantitative Comparison of Neuronal Segmentation Algorithms
| Algorithm | Primary Imaging Modality | Reported Accuracy/ Dice Coefficient | Strengths | Limitations | Computational Demand |
|---|---|---|---|---|---|
| Flood-Filling Networks (FFNs) | Electron Microscopy | N/R | Excels in segmenting long axons; handles large volumetric data | Easy detachment of thin spines from parent dendrites | Very high; process can take months |
| LSD-predicting U-Net | Electron Microscopy | N/R | Superior for segmenting myelinated axons | Same spine detachment issue as FFN | High |
| SENPAI | Confocal/STED Microscopy | Outperforms state-of-the-art tools | Accurate multi-scale segmentation from neurons to spines; handles densely packed cells | Performance depends on image quality from sample preparation | Medium |
| U-Net with Residual Units | Quantitative Phase Imaging (QPI) | 0.75 (when trained on augmented real data) | Effective for label-free imaging; compatible with dry mass calculations | Requires significant training data; poorer neurite segmentation | Medium |
| Cellpose | Fluorescence Microscopy | Widely adopted generalist model | Generalizable across cell types; user-friendly | Struggles with QPI-specific noise; cannot separate cytoplasm/nuclei in projections | Low to Medium |
N/R: Not reported in the analyzed literature
Specialized Segmentation Applications
For functional imaging of neuronal dynamics, a novel compressed acquisition method called DeMOSAIC has demonstrated remarkable capabilities for high-temporal-resolution imaging [73]. This approach optically segments predefined regions of interest (ROIs) and assigns each to a single pixel, enabling sampling rates exceeding 5 kHz—sufficient to resolve individual action potential waveforms within subcellular structures [73]. While not a segmentation algorithm per se, this hardware-based segmentation approach bypasses many computational challenges for applications requiring ultra-high temporal resolution.
Comparative Analysis of Feature Extraction Methodologies
Once segmentation is achieved, extracting meaningful morphological features becomes critical for classifying cell types and assessing cultural composition.
Performance Benchmarks for Feature Extraction
Table 2: Feature Extraction Performance in Mixed Neural Cultures
| Method | Cell Type Classification Accuracy | Mechanism of Action (MOA) Enrichment Score | Key Advantages | Key Limitations |
|---|---|---|---|---|
| Traditional Morphotextural Features (CellProfiler) | ~96% for neuroblastoma/astrocytoma lines [1] | Average: 62.6% across co-cultures [74] | Interpretable features; well-established pipeline | Hand-crafted features may miss subtle phenotypes |
| Pre-trained CNN (EfficientNetB0) | N/R | Range: 52.6-74.1% (mean: 61.0%) [74] | No need for domain-specific training; fast implementation | Variable performance across culture conditions |
| Pre-trained CNN (MobileNetV2) | N/R | Range: 52.6-74.1% (mean: 62.0%) [74] | Balanced performance and computational efficiency | Similar variability issues as EfficientNetB0 |
| CNN with Regional Restriction | >96% for iPSC-derived neural cultures [1] | N/R | Maintains high accuracy in dense cultures; reduces interference | Requires precise segmentation |
| Random Forest Classifier | ~75% F-score [1] | N/R | Simple implementation; works well with small datasets | Significantly lower accuracy compared to CNN approaches |
N/R: Not reported in the analyzed literature
Impact of Input Region on Feature Extraction
The choice of input region significantly affects classification performance in dense cultures. Research demonstrates that inputs containing the nuclear region of interest and its immediate environment achieve classification accuracy equally high as whole-cell inputs in semi-confluent cultures while maintaining prediction accuracy even in very dense cultures [1]. This regional restriction approach improves performance by reducing interference from neighboring cells in crowded environments.
Experimental Protocols for Method Validation
To ensure reproducible results, we provide detailed protocols for key experiments cited in our comparison.
Protocol for CNN-Based Cell Type Classification
This protocol is adapted from the unbiased identification of cell identity in dense mixed neural cultures [1]:
Cell Culture and Preparation:
- Culture mixed neural cultures under standard conditions. For validation, use defined co-cultures of neuroblastoma (SH-SY5Y) and astrocytoma (1321N1) cell lines.
- Plate cells at varying densities to test density-dependent effects.
Staining and Imaging:
- Implement a 4-channel confocal imaging protocol based on Cell Painting.
- Stain with appropriate fluorescent markers (e.g., Hoechst for nuclei, phalloidin for actin, etc.).
- Acquire images across multiple biological replicates.
Image Processing:
- Segment cells using a deep learning-based approach (e.g., Cellpose).
- Extract two sets of inputs for each cell: (a) whole-cell region, (b) nuclear region plus immediate surroundings.
Model Training and Validation:
- Implement a ResNet CNN architecture with appropriate pre-training.
- Train separate models on whole-cell and nuclear-focused inputs.
- Validate classification accuracy using stratified k-fold cross-validation.
- Compare performance against traditional random forest classifiers using hand-crafted morphological features.
Protocol for Evaluating Segmentation Quality
This protocol validates segmentation algorithms when manual ground truth is difficult to obtain [75]:
Sample Preparation:
- Use clarified brain tissues from L7GFP mouse line for improved image quality.
- Process samples for long-term immunochemistry.
Multi-Scale Imaging:
- Acquire correlative images using both low-magnification (20x) for tissue scanning and high-resolution (93x) confocal or 3D STED microscopy for spine-level detail.
- Implement tile imaging for wide mosaic brain areas with custom-made spacers to ensure sample flatness.
Algorithm Application:
- Apply SENPAI and comparison algorithms to the same datasets.
- For sparse labeling, use the G-cut meta-method for post-hoc separation of interweaving neurons.
Validation Without Full Manual Ground Truth:
- Compare morphometrics extracted from segmented neurons with established quantitative indices of Purkinje cell morphology from literature.
- Focus on metrics like dendritic length, branching complexity, and spine density.
- Evaluate detection and correct assignment of dendritic spines to parent neurons.
Workflow Visualization
The following diagram illustrates the integrated workflow for segmentation and feature extraction in dense neural cultures:
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents and Solutions for Neural Culture Analysis
| Reagent/Solution | Function | Example Application | Considerations |
|---|---|---|---|
| Basic Fibroblast Growth Factor (bFGF) | Promotes cortical neurogenesis; preserves neural progenitor population | Essential for long-term maintenance of primary human mixed brain cultures [72] | Critical difference from rodent cultures; significantly improves yield and survival |
| Cell Painting Dyes | Multiplexed staining of multiple cellular compartments | High-content morphological profiling; phenotypic screening [1] [76] | Enables unsupervised morphological profiling without predefined targets |
| Hoechst Stain | Nuclear counterstain | Cell segmentation and identification; cell counting [74] | Standard reference for cellular identification across modalities |
| CK8/18 Antibodies | Epithelial cell marker | Identifying cancer cells in co-culture systems [74] | Essential for specific cell type identification in heterogeneous cultures |
| Vimentin Antibodies | Mesenchymal cell marker | Identifying fibroblasts in co-culture systems [74] | Enables discrimination of different cell populations in mixed cultures |
| GFP-Labeled Cell Lines | Intrinsic neuronal labeling | Visualization of neuronal morphology in dense cultures [75] | Enables high-resolution imaging without antibody penetration issues |
| Refractive Index Matching Solutions | Tissue clearing | Deep imaging in clarified tissues for improved SNR and CNR [75] | Critical for super-resolution imaging of thick samples |
The comparative analysis presented in this guide demonstrates that method selection for segmenting and extracting features from dense, mixed neural cultures must be guided by specific research requirements and experimental constraints.
For segmentation tasks, the choice between FFNs, LSD U-Net, SENPAI, and generalist tools like Cellpose depends critically on imaging modality, required resolution, and cellular density. Similarly, feature extraction performance varies significantly between traditional handcrafted features and CNN-based approaches, with each showing distinct advantages in different culture conditions and applications.
When positioned within the broader thesis of morphological profiling versus immunocytochemistry for culture validation, these computational approaches offer compelling advantages in throughput, cost, and preservation of sample viability. However, immunocytochemistry remains essential for validating specific molecular identities and creating gold-standard references for training computational models.
As the field advances, we anticipate increased integration of these methodologies, with optimized computational pipelines providing rapid, non-destructive assessment of culture composition complemented by targeted molecular validation. This synergistic approach will ultimately accelerate research using complex neural culture systems across basic neuroscience and drug development applications.
Strategies for Managing High Heterogeneity in Organoid and Primary Culture Batches
The adoption of three-dimensional organoids and primary neural cultures has revolutionized neuroscience research, disease modeling, and drug development by providing more physiologically relevant human models. However, the transformative potential of these advanced cellular systems is constrained by a significant challenge: high batch-to-batch heterogeneity. This variability manifests in morphological, structural, and functional differences between individual organoids, even within the same differentiation batch, complicating experimental reproducibility and reliable data interpretation [77] [21]. The inherent stochastic nature of in vitro self-assembly and organ developmental selection drives this heterogeneity, leading to organoids with poorly controlled morphogenesis and varied cellular compositions [77]. In primary neural cultures, protocol variations and technical execution differences introduce similar variability, particularly affecting cellular composition, purity, and maturity, which directly influences gene expression and functional activity [1].
Within this context, two distinct methodological paradigms have emerged for validating culture composition and managing heterogeneity: traditional immunocytochemistry (ICC) and increasingly morphological profiling approaches. While ICC has long been the gold standard for cell type identification through specific biomarker detection, emerging morphological strategies leverage high-content imaging and computational analysis to provide rapid, non-destructive assessment of culture quality and composition [1] [21]. This guide objectively compares these approaches, providing experimental data and methodological details to help researchers select appropriate heterogeneity management strategies for their specific research contexts, particularly in neural culture validation.
Core Technologies for Culture Validation
Immunocytochemistry (ICC): The Established Standard
Immunocytochemistry remains the most widely adopted method for cell type identification and validation in neural cultures. This technique relies on antibodies conjugated to fluorescent tags to target specific intracellular and surface antigens, allowing researchers to characterize and quantify cellular composition through microscopy.
Experimental Protocol for Neural Culture ICC:
- Culture Fixation: Apply 4% paraformaldehyde in PBS for 15 minutes at room temperature
- Permeabilization and Blocking: Use 0.1% Triton X-100 with 5% normal serum from host species of secondary antibody for 30 minutes
- Primary Antibody Incubation: Apply species-specific validated antibodies (e.g., anti-MAP2 for neurons, anti-GFAP for astrocytes) diluted in blocking buffer overnight at 4°C
- Secondary Antibody Incubation: Apply fluorophore-conjugated secondary antibodies (e.g., Alexa Fluor varieties) for 1 hour at room temperature protected from light
- Counterstaining and Imaging: Use DAPI for nuclear staining and mount with anti-fade medium before imaging with epifluorescence or confocal microscopy [1] [21]
The strengths of ICC include high specificity for target antigens, ability to provide spatial distribution information within cultures, and well-established protocols with extensive antibody panels for neural cell types. However, limitations include the method's destructive nature preventing longitudinal studies, potential antibody cross-reactivity issues, low throughput capacity, and the inability to provide real-time monitoring of culture status [1].
Morphological Profiling: The Emerging Paradigm
Morphological profiling represents a innovative approach that utilizes quantitative analysis of cellular and structural features to identify cell types and assess culture quality without destructive processing. This methodology leverages high-content imaging combined with computational analysis, including machine learning algorithms, to classify cells and tissues based on their morphotextural fingerprints [1] [21].
Experimental Protocol for Cell Painting and Morphological Analysis:
- Sample Staining: Apply a panel of non-specific fluorescent dyes including:
- Hoechst 33342 or DAPI for nuclear staining
- Phalloidin for F-actin visualization (cytoskeleton)
- Concanavalin-A or Wheat Germ Agglutinin for glycoproteins/glycans
- SYTO 14 for RNA/nucleoli
- Mitotracker for mitochondria
- High-Content Imaging: Acquire multi-channel images using automated confocal or widefield microscopy systems
- Image Processing and Feature Extraction: Use segmentation algorithms to identify individual cells and subcellular compartments, then extract quantitative features describing shape, intensity, texture, and spatial relationships
- Machine Learning Classification: Train convolutional neural networks or random forest classifiers on extracted features to identify cell types and culture characteristics [1]
Advanced implementations of this approach have demonstrated remarkable classification accuracy. One study utilizing convolutional neural networks achieved above 96% accuracy in recognizing cell types in dense, mixed neural cultures, significantly outperforming traditional random forest classifiers [1]. The non-destructive nature of morphological profiling enables longitudinal monitoring of the same cultures over time, providing valuable insights into developmental trajectories and temporal dynamics of heterogeneity.
Comparative Experimental Data and Performance Metrics
Table 1: Quantitative Comparison of Validation Method Performance in Neural Cultures
| Performance Metric | Immunocytochemistry | Morphological Profiling |
|---|---|---|
| Classification Accuracy | >99% for validated targets (antigen-dependent) | 96-99% (method-dependent) [1] |
| Temporal Resolution | Endpoint measurements only | Real-time to longitudinal possible |
| Sample Preservation | Destructive | Non-destructive [21] |
| Throughput Capacity | Low to moderate | High to very high [1] |
| Multiplexing Capacity | Limited by spectral overlap | Highly multiplexed by design |
| Technical Variability | Antibody lot-dependent, user technique | Algorithm-dependent, consistent |
| Cost per Sample | Moderate to high (antibodies) | Low after initial setup |
Table 2: Experimental Validation of Morphological Selection for Cerebral Organoids
| Morphological Variant | Primary Cell Composition | Identification Marker | Selection Accuracy |
|---|---|---|---|
| Variant 1 (Rosette structures) | Cortical/glutamatergic neurons | EMX1, SLC17A7, NEUROD6 | >95% [21] |
| Variant 2 (Low transparency) | GABAergic neurons | GAD2, DLX1, DLX2, DLX5, DLX6 | >95% [21] |
| Variant 3/4 (Cystic/fibrous) | CNS fibroblasts | COL1A1 | >90% [21] |
| Variant 5 (Pigmented) | Melanocytes | TYR | >95% [21] |
| Variant 6/7 (Transparent/cystic) | Choroid plexus | TTR | >90% [21] |
Research has systematically correlated morphological features with cellular composition in cerebral organoids, demonstrating that visual characteristics reliably predict internal cellular makeup. Ikeda et al. paired single-cell RNA sequencing with morphological analysis to establish these correlations, enabling non-destructive identification of target cerebral cortical tissues with high accuracy [21]. This morphology-based selection approach significantly enhances experimental reproducibility and reliability for both research applications and cell-based therapies.
Integrated Experimental Workflows
Diagram 1: Integrated Culture Validation Workflow. This workflow combines morphological profiling for initial non-destructive quality control with targeted immunocytochemistry for specific validation and functional assays to assess physiological relevance.
Research Reagent Solutions for Culture Validation
Table 3: Essential Research Reagents for Culture Validation Methods
| Reagent Category | Specific Examples | Research Function | Application Context |
|---|---|---|---|
| Extracellular Matrix | Matrigel, Synthetic hydrogels (GelMA) | Provides 3D structural support for organoid growth | Organoid culture [77] [78] |
| ICC Primary Antibodies | Anti-EMX1 (cortical neurons), Anti-GAD2 (GABAergic neurons), Anti-COL1A1 (fibroblasts) | Cell type identification and validation | Target-specific validation [21] |
| Morphological Stains | Hoechst 33342, Phalloidin, Concanavalin-A, SYTO 14, Mitotracker | Non-specific labeling for feature extraction | Morphological profiling [1] |
| Stem Cell Maintenance | Wnt3A, R-spondin-1, Noggin, B27, N-2 | Maintains stemness and promotes differentiation | Organoid culture medium [78] |
| Neural Culture Supplements | Y-27632 (ROCK inhibitor), N-acetyl-l-cysteine | Enhances cell survival, reduces apoptosis | Primary neural culture [78] |
Technical Considerations and Implementation Strategy
Successful management of culture heterogeneity requires careful consideration of several technical factors. For organoid cultures, the source of extracellular matrix significantly influences reproducibility; Matrigel exhibits batch-to-batch variability, while synthetic hydrogels like GelMA provide more consistent chemical and physical properties [78]. In primary neural cultures, the choice between Banker-style "sandwich" cultures and mixed cultures represents a critical trade-off between experimental accessibility and model complexity, with Banker methods requiring greater technical expertise but providing superior conditions for single-cell imaging and analysis [38].
An effective implementation strategy should incorporate tiered validation approaches, beginning with rapid morphological assessment for batch quality control, followed by targeted immunocytochemistry on representative samples for specific marker validation, and culminating in functional characterization where appropriate. This balanced methodology maximizes throughput while maintaining analytical rigor. Furthermore, researchers should establish cell line-specific morphological benchmarks through initial comprehensive characterization, creating reference standards for subsequent batch evaluations.
The integration of automation technologies represents another powerful approach for reducing heterogeneity. Robotic liquid handling systems can perform precisely controlled tasks including initial stem cell allocation, media addition and replacement, and drug testing, significantly improving consistency across batches [77]. Similarly, high-throughput screening platforms enable comprehensive assessment of large organoid arrays, providing robust statistical power for heterogeneity analysis despite individual organoid variations.
Managing heterogeneity in organoid and primary culture batches remains a significant challenge in neuroscience research, but strategic implementation of complementary validation methodologies provides powerful solutions. Morphological profiling offers unprecedented advantages for non-destructive, high-throughput quality control, while immunocytochemistry continues to deliver essential specificity for target validation. The experimental data presented demonstrates that morphological approaches can achieve classification accuracy exceeding 96% in mixed neural cultures, establishing their reliability for routine batch assessment [1] [21].
Forward-looking research strategies will benefit from integrated workflows that leverage the respective strengths of both approaches, beginning with morphological screening for rapid batch qualification followed by targeted molecular validation. As these technologies evolve, particularly with advances in artificial intelligence integration and automated culture systems, researchers can anticipate increasingly sophisticated approaches for heterogeneity management that will enhance reproducibility across organoid and primary culture applications in basic research, drug development, and therapeutic implementation.
Implementing Non-Destructive, Label-Free Selection to Preserve Culture Viability
The field of neural culture validation is undergoing a significant transformation, moving from traditional, destructive analytical methods toward innovative non-destructive technologies. This shift is particularly crucial for applications in drug development and cell therapy, where preserving culture viability and sterility is paramount. Traditional validation methods, primarily immunocytochemistry and flow cytometry, require cell fixation, labeling with fluorescent antibodies, and disruptive sampling that ultimately destroy the cultured cells [79]. In contrast, emerging label-free technologies leverage advanced imaging and artificial intelligence to extract rich morphological and biophysical data without compromising culture integrity. This comparison guide objectively examines the performance of these competing approaches, providing researchers and drug development professionals with experimental data to inform their technology selection for neural culture applications.
Technology Comparison: Performance and Capabilities
Quantitative Performance Metrics
The table below summarizes key performance characteristics of label-free morphological profiling versus traditional immunocytochemistry for neural culture validation.
Table 1: Performance Comparison of Neural Culture Validation Methods
| Performance Parameter | Morphological Profiling | Immunocytochemistry |
|---|---|---|
| Classification Accuracy | 96% for cell types in mixed neural cultures [1] | >99% for specific protein targets |
| Culture Viability | Maintained at 100% (non-destructive) [79] | Lost (destructive endpoint assay) |
| Temporal Resolution | Continuous, real-time monitoring possible [79] | Single time point measurements |
| Multiplexing Capability | High (multiple morphological features simultaneously) | Limited by antibody spectral overlap |
| Assay Preparation Time | Minimal (direct imaging) | Extensive (fixation, permeabilization, labeling) |
| Sterility Maintenance | Preserved (closed-system integration) [79] | Compromised (sample removal required) |
Applications in Neural Culture Research
Table 2: Application-Specific Method Performance
| Research Application | Morphological Profiling Advantages | Immunocytochemistry Advantages |
|---|---|---|
| Neural Differentiation Monitoring | Non-destructive tracking of neurite outgrowth and morphological maturation [17] | Definitive protein expression confirmation (e.g., MAP2, Tau) [17] |
| Cell Type Identification | Unbiased identification in dense mixed cultures (96% accuracy) [1] | Specific marker-based identification (e.g., neurons, astrocytes, microglia) [1] |
| Functional State Assessment | Calcium dynamics correlated with differentiation state [17] | Direct receptor and synaptic protein quantification |
| High-Content Screening | Suitable for longitudinal studies and rare cell analysis [80] | Established protocols and extensive validated antibody libraries |
Experimental Protocols for Method Validation
Label-Free Morphological Profiling Protocol
Workflow Title: Neural Culture Profiling with qOBM and Live Imaging
Detailed Methodology:
Cell Culture Preparation: Plate SH-SY5Y neuroblastoma cells or induced pluripotent stem cell (iPSC)-derived neural cultures in appropriate vessels. For differentiation studies, treat with retinoic acid (10µM) for 6 days followed by brain-derived neurotrophic factor (BDNF, 50ng/ml) for an additional 6 days [17].
System Integration: Implement a compact quantitative oblique back-illumination microscopy (qOBM) system within the cell culture incubator (8×12×10 inches). Connect to bioreactor via closed-loop tubing with peristaltic pump for continuous monitoring [79].
Image Acquisition: Flow cells from bioreactor through imaging flow cell at acquisition rate of ~10 Hz. Capture quantitative phase images providing information on cellular dry mass and refractive index properties. For functional assessment, perform live calcium imaging using Fluo-4 (2µM) dye with confocal microscopy at 1 Hz sampling rate [17].
Data Processing: Extract morphological features using Cell Painting protocols with 4-6 channel imaging or deep learning approaches with convolutional neural networks (ResNet architecture). For calcium dynamics, analyze oscillation frequency, amplitude, and transient kinetics [1] [17].
Culture Maintenance: Return imaged cells to bioreactor for continued expansion, maintaining sterility and viability throughout the process [79].
Traditional Immunocytochemistry Protocol
Workflow Title: Endpoint Immunofluorescence Validation
Detailed Methodology:
Sample Extraction: Remove cells from culture vessel, disrupting the sterile environment and potentially affecting culture expansion, especially during early time points [79].
Fixation and Permeabilization: Wash cells with PBS and fix with 4% paraformaldehyde for 20 minutes. Permeabilize with 0.1% Triton X-100 for 10 minutes, then block with 2% FBS for 1 hour [17].
Antibody Labeling: Incubate with primary antibodies overnight at 4°C against neural markers (e.g., MAP2, Tau, PSD95, Synapsin). Wash and incubate with fluorescent secondary antibodies overnight at 4°C [17].
Image Acquisition: Capture images using confocal microscopy (e.g., ZEISS LSM 880) with appropriate laser lines (488 nm, 568 nm, 647 nm) and spatial resolution of 30 nm per pixel [17].
Quantitative Analysis: Process images to quantify protein expression levels, subcellular localization, and co-localization patterns. Use DAPI counterstain for nuclear identification.
Signaling Pathways in Neural Differentiation
Pathway Title: Calcium Dynamics in Neuronal Development
Pathway Description: Neuronal differentiation of SH-SY5Y cells involves distinct shifts in calcium dynamics that can be monitored non-invasively. Undifferentiated cells exhibit spontaneous high-amplitude slow calcium oscillations. Retinoic acid treatment initiates differentiation, triggering neurite outgrowth and expression of neuronal markers, while simultaneously abolishing spontaneous calcium oscillations. Subsequent BDNF treatment enhances neuronal polarization and enrichment with specific neuronal markers, accompanied by a resurgence of spontaneous calcium oscillations with faster kinetics. Carbachol stimulation in differentiated cells induces calcium transients with higher peaks and biphasic decay, indicating mature signaling capabilities. These functional changes provide measurable endpoints for non-destructive monitoring of differentiation status [17].
Research Reagent Solutions Toolkit
Table 3: Essential Materials for Non-Destructive Neural Culture Analysis
| Reagent/Resource | Function | Application Example |
|---|---|---|
| SH-SY5Y Cell Line | Human neuroblastoma model for neuronal differentiation studies | Sequential differentiation into morphologically mature neurons [17] |
| Retinoic Acid | Differentiation inducer, promotes neurite outgrowth | Used at 10µM concentration for 6 days to initiate neuronal differentiation [17] |
| Brain-Derived Neurotrophic Factor (BDNF) | Enhances neuronal maturation and survival | Applied at 50ng/ml for 6 days following RA treatment [17] |
| Fluo-4 AM Calcium Indicator | Fluorescent calcium sensor for functional imaging | Used at 2µM concentration to monitor spontaneous and evoked calcium dynamics [17] |
| qOBM Imaging System | Compact quantitative phase imaging for label-free monitoring | Enables continuous, non-destructive imaging within standard incubators [79] |
| Cell Painting Dyes | Multiplexed morphological profiling | Distinguishes cell types in mixed neural cultures with high accuracy [1] |
| Convolutional Neural Networks (CNN) | Deep learning for image analysis and classification | Achieves 96% accuracy in cell type identification [1] |
The experimental data presented in this comparison guide demonstrates that both morphological profiling and immunocytochemistry offer distinct advantages for neural culture validation. Label-free approaches provide unprecedented capabilities for continuous, non-destructive monitoring while preserving culture viability and sterility—critical considerations for manufacturing cell therapies and longitudinal studies. The 96% classification accuracy achieved through deep learning analysis of morphological data confirms the analytical robustness of these methods [1]. Traditional immunocytochemistry remains essential for definitive protein-specific validation and endpoint analysis. The optimal approach depends on specific research requirements: morphological profiling for longitudinal functional studies and therapeutic manufacturing, and immunocytochemistry for definitive marker expression confirmation. Future advancements will likely see increased integration of these complementary technologies, leveraging the strengths of each to accelerate drug development and cell therapy innovation.
Data Management and Standardization for High-Dimensional Morphological Datasets
In the field of neural culture validation research, the transition from traditional, targeted analytical methods to high-dimensional, unbiased profiling technologies has created both unprecedented opportunities and significant data management challenges. Morphological profiling, particularly through advanced assays like Cell Painting, generates immense datasets that capture subtle changes in cellular architecture resulting from genetic, chemical, or disease-state perturbations [81] [76]. Similarly, immunocytochemistry (ICC) remains a cornerstone technique for specific biomarker validation in neural cultures. The management, standardization, and integration of data from these complementary approaches represent a critical frontier in neuroscience research and drug development.
The complexity of these high-content datasets necessitates sophisticated data management frameworks that can handle not only the volume but also the multidimensional nature of the information. Where ICC provides focused, hypothesis-driven insights into specific neural markers, morphological profiling offers a systems-level, discovery-oriented perspective on cellular state [82] [83]. This comparison guide examines the data management requirements, standardization approaches, and performance characteristics of these methodologies within the specific context of neural culture validation, providing researchers with a practical framework for selecting and implementing appropriate data management strategies for their experimental needs.
Comparative Performance Analysis: Morphological Profiling vs. Immunocytochemistry
Technical Performance and Applications
Table 1: Technical and performance comparison between morphological profiling and immunocytochemistry for neural culture validation.
| Parameter | Morphological Profiling (Cell Painting) | Immunocytochemistry (ICC) |
|---|---|---|
| Primary Application | Unbiased discovery, mechanism of action studies [81] [83] | Targeted validation of specific biomarkers [82] |
| Multiplexing Capacity | High (6-8 cellular components simultaneously) [81] | Moderate (typically 2-4 targets with spectral overlap) |
| Throughput Potential | High (suitable for large-scale screening) [1] [76] | Low to moderate (limited by antibody incubation) |
| Quantitative Output | ~1,000 morphological features [81] | Semi-quantitative intensity measurements [82] |
| Data Density | Extremely high (features × samples × conditions) [81] | Moderate (limited to targeted markers) |
| Standardization Challenge | Feature extraction reproducibility, batch effects [76] | Antibody validation, staining consistency [82] |
| Information Content | System-level morphological responses [1] [83] | Specific protein localization and expression [82] |
Experimental Validation Metrics
Table 2: Experimental performance metrics for neural culture validation applications.
| Performance Metric | Morphological Profiling | Immunocytochemistry |
|---|---|---|
| Cell Type Classification Accuracy | >96% for neural lineages [1] | >98% for specific markers (e.g., ER/PR) [82] |
| Concordance with Transcriptomics | Moderate (shared subspace) [81] | Not directly comparable |
| Technical Reproducibility | High (multi-site correlation >0.8) [76] | Moderate (kappa 0.89-0.98) [82] |
| Temporal Resolution | Fixed timepoints (hours to days) | Fixed timepoints (hours to days) |
| Scalability | Highly scalable for compound screening [76] [83] | Limited by antibody costs and processing time |
| Cost per Sample | Moderate (reagents and imaging) [1] | Variable (antibody-dependent) |
Experimental Protocols and Methodologies
Cell Painting Assay Protocol for Neural Cultures
The Cell Painting assay provides a comprehensive workflow for capturing high-dimensional morphological data from neural cultures [81] [1]. The following protocol has been specifically adapted for neural cell types, including iPSC-derived neurons, astrocytes, and mixed neural cultures:
Cell Culture and Plating: Plate neural cells in 96-well or 384-well imaging plates at optimized densities for neural cultures (typically 5,000-15,000 cells/well for 96-well format). For iPSC-derived neural cultures, ensure consistent maturity and differentiation status across plates [1].
Staining Protocol:
- Fixation: 4% formaldehyde for 20 minutes at room temperature
- Permeabilization: 0.1% Triton X-100 for 15 minutes
- Staining cocktail incubation for 30 minutes containing:
- Mitotracker Red CMXRos (mitochondria)
- Phalloidin (actin cytoskeleton)
- Wheat Germ Agglutinin (plasma membrane)
- Concanavalin A (endoplasmic reticulum)
- Hoechst 33342 (nucleus)
- Wash steps with PBS between each staining reagent [81]
Image Acquisition:
- Acquire images using high-content screening microscopes with 20x or 40x objectives
- Capture 5-9 fields per well to ensure adequate cell sampling
- Image each fluorescent channel separately with appropriate filter sets [1]
Image Analysis and Feature Extraction:
Immunocytochemistry Protocol for Neural Marker Validation
For targeted validation of specific neural cell types, ICC provides a complementary approach with high specificity for established biomarkers:
Sample Preparation:
- Culture neural cells on coverslips or in imaging plates
- Fix with 4% paraformaldehyde for 15 minutes
- Permeabilize with 0.3% Triton X-100 for 10 minutes [82]
Antibody Staining:
- Block with 5% normal serum for 1 hour
- Incubate with primary antibodies for 2 hours at room temperature or overnight at 4°C
- Common neural markers: β-III-tubulin (neurons), GFAP (astrocytes), IBA1 (microglia)
- Wash thoroughly with PBS
- Incubate with fluorophore-conjugated secondary antibodies for 1 hour
- Counterstain with DAPI for nuclear visualization [82] [1]
Image Acquisition and Analysis:
- Acquire images using epifluorescence or confocal microscopy
- Quantify signal intensity and cellular localization
- For receptor staining (e.g., ER/PR), use established scoring systems (Allred score) [82]
Data Management Workflows and Standardization Approaches
Comparative Data Management Pipelines
Diagram 1: Comparative data management workflows for morphological profiling and immunocytochemistry.
Data Standardization Challenges and Solutions
Morphological Profiling Standardization
The high-dimensional nature of morphological profiling data presents unique standardization challenges. Effective management requires addressing several critical aspects:
Batch Effect Correction: Implementing robust normalization methods to account for technical variation across experimental batches, plates, and imaging sessions [76]. Multi-site studies have demonstrated that with extensive optimization, correlation coefficients >0.8 can be achieved across different laboratories [76].
Feature Standardization: Developing consistent feature extraction protocols across different Cell Painting implementations. This includes standardizing CellProfiler pipelines or alternative segmentation approaches to ensure comparable feature sets [81] [1].
Metadata Annotation: Comprehensive metadata capture following FAIR (Findable, Accessible, Interoperable, Reusable) data principles. Critical metadata includes experimental conditions, cell type specifications, staining protocols, imaging parameters, and analysis workflows [81].
Immunocytochemistry Standardization
While ICC generates less complex data, standardization remains essential for reproducible results:
Antibody Validation: Rigorous validation of antibody specificity using appropriate controls, including knockdown/knockout validation where possible [82].
Quantification Standards: Implementation of consistent scoring systems, such as the Allred score for hormone receptors or established thresholds for neural marker classification [82] [1].
Cross-platform Reproducibility: Standardization of staining protocols, image acquisition settings, and analysis methods to enable data comparison across experiments and laboratories [82].
Integration with Complementary Data Modalities
Multi-modal Data Integration Framework
Diagram 2: Multi-modal data integration framework for neural culture validation.
Cross-Modal Prediction and Validation
The integration of morphological profiling with complementary data types enables powerful applications in neural culture validation:
Shared Subspace Utilization: Research has demonstrated that morphological profiles and gene expression data share a common subspace that enables cross-modal prediction. Machine learning models can predict approximately 30-40% of gene expression features from morphological profiles alone, with multilayer perceptrons outperforming linear models for this task [81].
Complementary Information Leveraging: Each modality contains both shared and unique information. While the shared subspace enables prediction across modalities, the modality-specific information provides complementary insights. For example, morphological profiling can capture post-translational modifications and cellular organization features not evident in transcriptomic data [81].
Validation Paradigms: ICC serves as a crucial bridging technology between high-content morphological profiling and molecular analyses. Targeted ICC validation of specific neural markers (e.g., β-III-tubulin for neurons, GFAP for astrocytes) provides ground truth data for interpreting morphological profiles in mixed neural cultures [82] [1].
Essential Research Reagent Solutions
Table 3: Key research reagents and computational tools for high-dimensional morphological data generation and analysis.
| Category | Specific Solution | Application in Neural Cultures | Performance Considerations |
|---|---|---|---|
| Cell Staining | Cell Painting Kit | Comprehensive morphological profiling of neural cells | Standardized staining for reproducibility [81] |
| Antibodies | Neural Marker Panels (β-III-tubulin, GFAP, etc.) | Specific cell type identification and validation | Requires rigorous validation [82] [1] |
| Image Analysis | CellProfiler | Feature extraction from neural images | Open-source, customizable pipelines [81] [1] |
| Data Integration | Cross-modal Autoencoders | Integrating morphology with transcriptomics | Learns shared latent space [81] |
| Quality Control | PCA/MDS Visualization | Batch effect detection and data quality assessment | Identifies technical artifacts [76] |
| Classification | Convolutional Neural Networks | Cell type identification in mixed cultures | >96% accuracy for neural lines [1] |
| Feature Selection | Morphological Subprofile Analysis | Identifying MoA-specific feature sets | Enables bioactivity annotation [83] |
The effective management and standardization of high-dimensional morphological datasets require thoughtful consideration of research objectives, technical capabilities, and analytical resources. Morphological profiling offers unparalleled discovery potential for characterizing novel neural culture responses, compound mechanisms of action, and system-level cellular changes, while immunocytochemistry provides targeted, specific validation of key neural markers with established clinical relevance.
For researchers implementing these technologies, a tiered approach is often most effective: utilizing morphological profiling for initial discovery and hypothesis generation in neural culture characterization, followed by targeted ICC validation of key findings using specific neural markers. The integration of these complementary approaches, supported by robust data management practices and standardized protocols, enables comprehensive neural culture validation that leverages both the unbiased nature of high-content morphological profiling and the specificity of immunocytochemistry.
As the field advances, increased standardization of data formats, analysis pipelines, and validation metrics will further enhance the reproducibility and translational potential of both morphological profiling and immunocytochemistry in neural culture research. By adopting the data management frameworks and comparative approaches outlined in this guide, researchers can effectively navigate the complexities of high-dimensional morphological data while generating biologically meaningful insights into neural culture composition and function.
In neural culture research, ensuring the accurate identity and composition of cellular populations is fundamental to experimental validity. Two principal methodologies have emerged for this validation: morphological profiling, a label-free approach that quantifies cellular shape and texture, and immunocytochemistry (ICC), a molecular technique that detects specific protein markers using antibodies [1] [84]. This guide provides an objective comparison of these two approaches, framing them not as competitors but as complementary tools for cross-validation. The central challenge in neural cultures, particularly those derived from induced pluripotent stem cells (iPSCs), is their inherent heterogeneity and the potential for variable differentiation outcomes [1]. Researchers must therefore employ robust validation strategies to confirm the presence and proportion of target cell types, such as neurons, astrocytes, and microglia. While ICC has long been the established standard, offering high molecular specificity, advances in machine learning and high-content imaging are making quantitative morphological profiling an increasingly powerful and efficient alternative [1] [85]. This guide will compare the performance, experimental requirements, and practical applications of both methods, providing researchers with the data needed to design rigorous culture validation protocols.
Methodological Comparison: Principles and Protocols
Immunocytochemistry (ICC): The Molecular Standard
ICC is an antibody-based technique that allows for the detection and localization of specific antigens within cells. The core principle relies on the specific binding of a primary antibody to a target protein, which is then visualized using a labeled secondary antibody or a directly conjugated primary antibody [86] [84]. The standard protocol involves several critical steps: cell fixation to preserve morphology and immobilize antigens, membrane permeabilization to allow antibody penetration, blocking to reduce non-specific binding, incubation with primary and then fluorescently-labeled secondary antibodies, and finally, counterstaining with dyes like DAPI to visualize nuclei [86]. The result is a highly specific, often multiplexed visualization of target proteins, which can be analyzed qualitatively or quantitatively via fluorescence microscopy.
Morphological Profiling: The Computational Alternative
Morphological profiling, often implemented through a method called Cell Painting, uses high-content imaging and computational analysis to identify cell types based on their inherent morphotextural fingerprints [1]. Instead of targeting specific proteins, this approach uses a panel of non-specific fluorescent dyes to label various cellular compartments such as the nucleus, endoplasmic reticulum, and Golgi apparatus. The resulting images are then processed using machine learning algorithms, such as Convolutional Neural Networks (CNNs), which extract thousands of morphological features related to cell shape, size, texture, and intensity [1]. These features form a unique profile that can be used to classify cell types with high accuracy, even in dense, mixed cultures, without the need for specific molecular labels.
Experimental Performance and Validation Data
Direct comparisons in research settings have quantified the relative strengths of both methods. The following table summarizes key performance metrics from published studies.
Table 1: Comparative Performance of Morphological Profiling and ICC
| Metric | Morphological Profiling | Immunocytochemistry (ICC) |
|---|---|---|
| Reported Classification Accuracy | >96% for neuroblastoma/astrocytoma cell lines [1] | ~99% purity for microglia in tertiary cultures (reference standard) [87] |
| Typical Purity in Mixed Neural Cultures | 96% vs. 86% population-level prediction for iPSC-derived neurons [1] | 73.3% ± 17.8% in secondary, 93.1% ± 6.0% in tertiary cultures [87] |
| Key Advantage | Label-free, non-destructive, single-cell resolution in mixed cultures [1] | High molecular specificity, direct antigen confirmation [84] [87] |
| Primary Limitation | Relies on trained models; indirect inference of identity [1] | Destructive, requires specific antibodies, lower throughput [1] [87] |
A pivotal study highlighted the power of morphological profiling, demonstrating that a CNN-based classifier could distinguish between neural cell lines with an accuracy exceeding 96% [1]. Furthermore, when applied to iPSC-derived neuronal cultures, a cell-based morphological prediction significantly outperformed a simple population-level assessment (96% vs. 86% accuracy) for determining the ratio of postmitotic neurons to neural progenitors [1]. In contrast, ICC remains the benchmark for ultimate purity validation. Quantitative analyses show that while secondary microglia-enriched cultures achieved about 73% purity, subsequent tertiary cultures could consistently reach purities of 93% or higher, with one benchmark reaching ~99% purity for microglia [87]. This demonstrates ICC's role as a definitive validation tool, albeit often after multi-step, low-yield processes.
The Scientist's Toolkit: Essential Reagents and Materials
Successful implementation of either validation strategy requires specific reagents. The following table details essential items for both methodologies.
Table 2: Essential Research Reagents for Culture Validation
| Item | Function/Description | Primary Method |
|---|---|---|
| Primary Antibodies | Bind specifically to target antigens (e.g., Iba1 for microglia, GFAP for astrocytes) [87]. | ICC |
| Fluorophore-Labeled Secondary Antibodies | Bind to primary antibodies to generate a detectable signal (e.g., Alexa Fluor dyes) [86]. | ICC |
| Cell Painting Dye Panel | A set of fluorescent dyes that non-specifically label key cellular organelles [1]. | Morphological Profiling |
| Blocking Agent (e.g., BSA, Serum) | Reduces non-specific antibody binding to minimize background signal [86] [88]. | ICC |
| Permeabilization Agent (e.g., Digitonin, Triton X-100) | Allows antibodies to cross cell and nuclear membranes by creating pores [86]. | ICC |
| Fixative (e.g., Paraformaldehyde) | Preserves cellular morphology and immobilizes antigens for analysis [86] [88]. | ICC & Morphological Profiling |
| Mounting Medium with DAPI | Preserves samples and provides a nuclear counterstain for spatial reference [86]. | ICC & Morphological Profiling |
Experimental Workflow and Signaling Pathways
The journey from cell culture to validated data involves distinct workflows for each method. The diagrams below illustrate the key steps and decision points for both ICC and morphological profiling.
Immunocytochemistry Workflow
ICC Experimental Workflow
Morphological Profiling Workflow
Morphological Profiling Workflow
Integrated Cross-Validation Strategy
For the highest confidence in neural culture validation, an integrated approach that leverages the strengths of both methods is recommended. A powerful strategy involves using morphological profiling as a high-throughput, non-destructive screening tool to monitor culture composition and health in real-time. This can be used to identify optimal time points for more detailed analysis or to quickly flag batches of cultures with atypical phenotypes. Subsequently, ICC can be employed as a definitive, low-throughput confirmatory test on a subset of samples to validate the findings from the morphological profile and provide explicit molecular identification [1] [87]. This cross-validation framework is particularly valuable for quality control in iPSC-derived neural cultures, where it can help standardize outcomes across different cell lines and differentiation batches, ultimately improving reproducibility in downstream applications like drug screening and disease modeling [1] [85].
The Validation Benchmark: A Direct Comparison of Accuracy, Efficiency, and Applicability
The rising utilization of complex in vitro neural models, including induced pluripotent stem cell (iPSC)-derived neurons, mixed neural cultures, and cerebral organoids, has created an urgent need for robust, scalable quality control methods. [1] [62] Traditional validation approaches like immunocytochemistry (ICC) provide specific molecular information but face significant limitations in throughput, cost, and destructiveness. In response, technological advances have catalyzed the development of morphological profiling as a powerful alternative for cell identity assessment. This guide provides an objective comparison between these competing methodologies, presenting quantitative performance data, experimental protocols, and cost-benefit analysis to inform research and drug development workflows.
The variability inherent in iPSC-derived neural cultures directly impacts experimental reproducibility and therapeutic development. As noted in recent studies, "genetic drift, clonal and patient heterogeneity cause variability in reprogramming and differentiation efficiency," hindering the adoption of these models in systematic drug screening pipelines. [1] Current validation methods, including combinations of sequencing, flow cytometry, and immunocytochemistry, are often "low in throughput, costly and/or, destructive," creating bottlenecks in research and development timelines. [1]
Performance Comparison: Morphological Profiling vs. Immunocytochemistry
The table below summarizes direct performance comparisons between morphological profiling and immunocytochemistry across key metrics relevant to neural culture validation:
Table 1: Performance Comparison of Neural Culture Validation Methods
| Performance Metric | Morphological Profiling | Traditional Immunocytochemistry |
|---|---|---|
| Classification Accuracy | Up to 96-97.5% (CNN-based) [1] | >99% (antibody-dependent) |
| Multiplexing Capability | 4-6 channels simultaneously [1] | Typically 2-4 antigens per experiment |
| Throughput Potential | High (automated, 96/384-well plates) [32] | Low to moderate (manual processing) |
| Temporal Resolution | Real-time possible (live-cell imaging) [32] | Endpoint measurements only |
| Cost per Sample | Low (after initial setup) [1] | High (reagent costs accumulate) |
| Destructive to Sample | No (compatible with live cells) [32] | Yes (requires fixation) |
| Assay Development Time | Weeks to months (algorithm training) | Days to weeks (antibody validation) |
Key Performance Differentiators
- Accuracy: Morphological profiling using convolutional neural networks (CNNs) has demonstrated exceptional accuracy in discriminating neural cell types, achieving above 96% classification fidelity in dense, mixed neural cultures, rivaling traditional methods. [1]
- Throughput: Live-cell imaging systems enable continuous monitoring of neuronal development without fixation, allowing real-time analysis of neurite outgrowth and network formation over days or weeks. [32]
- Cost-Benefit: While requiring substantial initial investment in imaging infrastructure and computational resources, morphological profiling offers significantly lower per-sample costs and reduces reagent expenses by eliminating the need for expensive antibodies for routine quality control. [1]
Experimental Protocols and Methodologies
High-Accuracy Morphological Profiling Protocol
The following workflow details the experimental protocol for implementing high-accuracy morphological profiling, based on methodologies that achieved up to 97.5% classification accuracy: [1]
Table 2: Key Research Reagent Solutions for Morphological Profiling
| Reagent/Resource | Function/Purpose | Example Specifications |
|---|---|---|
| Cell Painting Dyes | Multiplexed morphological staining | 4-6 channel fluorescence (DNA, RNA, protein, etc.) [1] |
| High-Content Imager | Automated image acquisition | Confocal microscopy, 40x objective or higher [1] |
| Convolutional Neural Network | Image analysis and classification | ResNet architecture [1] |
| Specialized Cell Culture Media | Maintain neuronal viability and function | Serum-free formulations with neurotrophic factors [89] |
| Live-Cell Imaging Chamber | Environmental control during imaging | Maintains 37°C, 5% CO₂ [32] |
Workflow Steps:
Sample Preparation: Plate neural cultures in appropriate density on imaging-compatible plates. For iPSC-derived cultures, ensure proper differentiation timing. [1]
Staining Protocol: Implement a modified Cell Painting assay using a 4-channel confocal imaging approach:
- Channel 1: Nuclear stain (e.g., Hoechst)
- Channel 2: Cytoplasmic stain (e.g., Phalloidin)
- Channel 3: RNA stain (e.g., SYTO RNA)
- Channel 4: Mitochondrial or other organelle stain [1]
Image Acquisition: Acquire images using high-content microscopy systems. For dense cultures, focus on the nuclear region of interest and its immediate environment, which maintains high prediction accuracy even in confluent conditions. [1]
Image Analysis:
- Use convolutional neural networks (CNNs) rather than traditional feature extraction methods
- Implement ResNet architecture for optimal performance
- Train models with approximately 5,000 training instances per cell class for optimal accuracy [1]
Validation: Compare classification results with a subset of immunocytochemistry validation samples to confirm accuracy.
Figure 1: Morphological profiling workflow for neural culture validation
Traditional Immunocytochemistry Validation Protocol
Workflow Steps:
Sample Fixation: Fix cells with 4% paraformaldehyde for 15-20 minutes at room temperature.
Permeabilization and Blocking: Permeabilize with 0.1% Triton X-100 for 10 minutes, followed by blocking with 2-5% serum for 1 hour.
Antibody Incubation:
- Primary antibody incubation overnight at 4°C (e.g., MAP2 for neurons, GFAP for astrocytes)
- Secondary antibody incubation for 1-2 hours at room temperature
Image Acquisition and Analysis: Acquire images using fluorescence microscopy and perform manual or semi-automated cell counting. [17]
Technical Mechanisms: How Morphological Profiling Achieves High Accuracy
Computational Architecture Advantages
Morphological profiling leverages deep learning architectures to achieve superior classification performance compared to traditional machine learning approaches:
- CNN Superiority: Convolutional Neural Networks significantly outperform random forest classifiers (F-score: 0.96 vs. 0.75) in cell type classification, due to their ability to automatically learn relevant features without manual feature engineering. [1]
- Region-Based Analysis: Iterative data erosion demonstrates that inputs containing the nuclear region and immediate environment achieve equally high classification accuracy as whole-cell inputs, preserving prediction accuracy even in dense cultures. [1]
- Feature Learning: CNNs excel at identifying subtle morphotextural fingerprints that distinguish neural cell types, including both texture features (e.g., Nucleus Channel 3 Energy) and shape metrics (e.g., Cellular Area). [1]
Figure 2: CNN architecture for neural cell classification
Application-Specific Implementation
Quality Control Framework for Neural Cultures
Implementing a tiered quality control framework maximizes efficiency while maintaining accuracy:
- Initial QC: Use non-invasive morphological assessments (organoid morphology, size, growth profile) for preliminary screening. [62]
- Final QC: Apply comprehensive analysis including cellular composition, cytoarchitectural organization, and cytotoxicity for validated cultures. [62]
- Live-Cell Integration: Incorporate real-time analysis systems (e.g., IncuCyte) for continuous monitoring of neurite outgrowth and network development without fixation. [32]
Specialized Applications
- Dense Mixed Cultures: Regional restriction to nuclear and peri-nuclear areas maintains classification accuracy above 96% even in confluent conditions. [1]
- Microglia Discrimination: A tiered strategy allows unequivocal discrimination of microglia from neurons, with further distinction between activated and non-activated states. [1]
- Organoid Validation: Standardized scoring systems evaluating morphology, size, cellular composition, cytoarchitectural organization, and cytotoxicity enable objective quality assessment. [62]
Cost-Benefit Analysis and Implementation Strategy
The table below outlines a comprehensive cost-benefit analysis to guide selection between validation approaches:
Table 3: Comprehensive Cost-Benefit Analysis of Validation Methods
| Consideration | Morphological Profiling | Immunocytochemistry |
|---|---|---|
| Initial Investment | High (imaging systems, computing) | Moderate (microscopy) |
| Recurring Costs | Low (dyes, media) | High (antibodies, reagents) |
| Personnel Requirements | Computational biology expertise | Cell biology techniques |
| Scalability | Excellent (high-throughput compatible) | Limited (labor-intensive) |
| Data Richness | High (multiparametric) | Targeted (specific markers) |
| Regulatory Acceptance | Emerging | Well-established |
| Adaptability to New Targets | Requires retraining | New antibody validation |
Strategic Implementation Recommendations
Large-Scale Screening: Morphological profiling offers superior cost-efficiency and throughput for drug screening applications and routine quality control of iPSC-derived neural cultures. [1]
Targeted Validation: Immunocytochemistry remains valuable for hypothesis-driven research requiring specific molecular marker confirmation and regulatory submissions.
Hybrid Approach: Implement morphological profiling for routine QC and initial screening, with ICC confirmation for critical endpoints, optimizing both efficiency and specificity.
Morphological profiling represents a transformative approach for neural culture validation, offering exceptional classification accuracy (up to 97.5%), significantly enhanced throughput, and favorable long-term cost-benefit compared to traditional immunocytochemistry. While ICC maintains advantages for specific molecular validation, the demonstrated performance of CNN-based morphological analysis establishes it as a powerful tool for researchers and drug development professionals working with complex neural models. As the field advances toward more standardized quality control frameworks, integrating these complementary technologies will maximize both efficiency and reliability in neuroscience research and therapeutic development.
The reproducibility of cerebral organoid cultures is paramount for their application in disease modeling, drug screening, and regenerative medicine. A significant challenge in the field is the inherent organoid-to-organoid variation, which persists even with standardized differentiation protocols [90]. This variability necessitates robust quality control methods to ensure the accurate and reliable collection of desired organoid types for downstream applications. Two principal approaches have emerged for validating neural culture composition: non-destructive morphological profiling and high-resolution molecular analysis via single-cell RNA sequencing (scRNA-seq). This guide provides an objective comparison of these two methodologies, framing them within the broader context of morphological profiling versus molecular techniques for neural culture validation. It is designed to inform researchers, scientists, and drug development professionals in selecting the appropriate validation strategy for their experimental and translational goals.
The two classification methods represent fundamentally different approaches, from sample processing to data output. Morphology-based classification relies on high-content imaging and computational analysis of physical characteristics, offering a non-destructive path to quality assessment [90] [1]. In contrast, scRNA-seq classification provides a high-resolution, molecular snapshot of cellular heterogeneity at the cost of being an end-point assay [90] [91]. The following diagram illustrates the core workflows for each method, highlighting their divergent paths and the potential for integration.
Performance Comparison and Experimental Data
A direct, paired comparison of these methodologies was performed in a recent study, which validated non-destructive morphology-based selection of cerebral cortical organoids by pairing it with scRNA-seq analysis [90]. The key findings and comparative performance metrics are summarized in the table below.
Table 1: Quantitative Comparison of Classification Method Performance
| Performance Metric | Morphology-Based Classification | scRNA-seq Classification |
|---|---|---|
| Classification Accuracy | High accuracy in distinguishing cerebral cortical tissues from other cerebral tissues [90] | Serves as the gold standard for definitive cell type identification [90] [91] |
| Primary Output | Morphological classes (e.g., "Cortical-like," "Non-neuronal") [90] | Detailed cell type composition (e.g., progenitors, glutamatergic neurons, astrocytes) [91] |
| Key Advantage | Non-destructive; enables selective collection of desired organoids for further culture/experimentation [90] | Unbiased, comprehensive profiling of cellular heterogeneity and novel states [91] |
| Identified Cell Types | Correlated with major tissue classes (neural crest, choroid plexus) [90] | Resolves all major neural lineages and subpopulations; maps to developing human brain atlas [91] |
| Best Use Case | High-throughput, cost-effective quality control and selection for ongoing experiments [90] [1] | In-depth characterization of model fidelity, protocol development, and disease mechanism investigation [91] |
The integrated transcriptomic Human Neural Organoid Cell Atlas (HNOCA), which encompasses over 1.7 million cells from 26 distinct protocols, exemplifies the power of scRNA-seq for benchmarking. It allows for the quantitative assessment of which primary brain cell types are generated—or are under-represented—in a given organoid protocol [91]. For instance, while telencephalic cell types are well-represented, certain thalamic, midbrain, and cerebellar neurons (e.g., Purkinje cells) remain scarce [91].
Detailed Experimental Protocols
To facilitate the adoption of these techniques, below are detailed protocols for the key experiments cited in this comparison.
Protocol: Morphology-Based Classification and Selection
This protocol is adapted from the paired morphological and scRNA-seq validation study [90] and enhanced by high-content morphological profiling principles [1].
Imaging and Staining (Optional):
- Culture cerebral organoids using established differentiation protocols.
- For high-content profiling, stained with a simplified, non-toxic Cell Painting dye kit (e.g., labelling nuclei, cytoplasm, and cytoskeleton) [1].
- Acquire high-resolution images using a confocal microscope. For label-free selection, bright-field imaging may suffice [90].
Morphological Feature Extraction:
- Use automated image analysis software (e.g., CellProfiler) to segment individual organoids or cells.
- Extract hundreds of quantitative morphological features describing shape, texture, and intensity. These can include "Cellular Area," "Nucleus Contrast," and other textural metrics [1].
Machine Learning Classification:
- Train a convolutional neural network (CNN) classifier on a curated set of images with known organoid subtypes or cellular compositions [1]. CNNs have been shown to outperform traditional random forest classifiers in this context, achieving accuracies above 96% in mixed neural cultures [1].
- Apply the trained model to new, unlabeled organoid images to predict their class (e.g., "cerebral cortical" vs "other") [90].
Selection:
- Based on the morphological classification, manually or robotically select organoids with the desired morphological features for subsequent experiments, ensuring they are not damaged in the process [90].
Protocol: scRNA-seq for Cell Type Validation
This protocol outlines the core steps for using scRNA-seq to validate organoid composition, as utilized in building the HNOCA [91] and paired validation studies [90].
Sample Preparation and Single-Cell Dissociation:
- Pool multiple organoids per condition or protocol to capture inherent variability.
- Dissociate organoids into a single-cell suspension using enzymatic (e.g., Trypsin/EDTA) and mechanical trituration methods [92].
- Resuspend cells in a suitable buffer (e.g., PBS with low BSA) and filter through a flow cytometry strainer to remove clumps.
Library Preparation and Sequencing:
- Process the single-cell suspension using a droplet-based platform (e.g., 10x Genomics) according to the manufacturer's instructions.
- This step captures individual cells, lyses them, and barcodes the resulting mRNA.
- Construct sequencing libraries and sequence on an Illumina platform to a sufficient depth (e.g., 50,000 reads per cell).
Bioinformatic Analysis and Atlas Mapping:
- Preprocessing: Use pipelines (e.g., CellRanger) to demultiplex data, align reads to a reference genome, and generate a gene-cell count matrix.
- Integration and Clustering: Perform quality control, normalization, and integration using tools like scPoli to correct for batch effects [91]. Cluster cells based on transcriptional similarity.
- Cell Type Annotation: Identify cluster-specific marker genes. Annotate cell types by mapping clusters to a reference atlas, such as the developing human brain [91], using label-transfer methods. Tools like RSS can estimate similarity to primary cell types [91].
- Analysis: Calculate presence scores for primary cell types to evaluate protocol fidelity and identify under-represented populations [91].
The Scientist's Toolkit: Essential Research Reagents and Materials
The following table details key reagents and tools essential for implementing the described classification methodologies.
Table 2: Essential Research Reagent Solutions for Organoid Classification
| Item | Function/Application | Specific Examples / Notes |
|---|---|---|
| Human Pluripotent Stem Cells (iPSCs) | Starting material for generating all cerebral organoids. | Use characterized, karyotypically normal lines (e.g., integration-free Sendai virus reprogrammed iPSCs) [93]. |
| Neural Differentiation Media | Supports the differentiation of iPSCs into cerebral organoids. | Often includes supplements like B-27 Plus and CultureOne to control glial expansion [92]. |
| Cell Painting Dye Kit | Fluorescent dyes for high-content morphological profiling. | Typically a 4-6 dye combination staining nuclei, cytoplasm, mitochondria, Golgi, and cytoskeleton [1]. |
| Single-Cell RNA Sequencing Kit | For generating barcoded scRNA-seq libraries from dissociated cells. | Droplet-based kits (e.g., 10x Genomics Chromium) are widely used [90] [91]. |
| Antibodies for Cell Type Validation | Confirm the presence of specific neural cell types via immunostaining. | Key markers: SOX2 (progenitors), TBR1/CTIP2/SATB2 (cortical neurons), GFAP (astrocytes), MBP (oligodendrocytes) [94]. |
| Reference Transcriptomic Atlas | Gold-standard dataset for annotating and benchmarking organoid cell types. | The Human Neural Organoid Cell Atlas (HNOCA) or developing human brain atlases are critical resources [91]. |
Integrated Analysis and Application Context
The choice between morphological and scRNA-seq classification is not mutually exclusive; rather, the methods are powerfully synergistic. The ultimate validation of a morphology-based approach comes from paired scRNA-seq analysis, which confirms that distinct morphological classes indeed correspond to unique cellular compositions [90]. Once this correlation is established, morphology becomes a powerful, standalone tool for rapid, non-destructive quality control.
The application context is a primary driver for method selection. Morphological profiling is ideal for high-throughput settings, such as quality control for cell-based therapies or large-scale drug screening, where preserving the biological material is essential [90] [1]. Conversely, scRNA-seq is indispensable for foundational research, including protocol development and optimization, where understanding the precise cellular outcomes and benchmarking against primary human data is crucial [91] [94]. It is also the preferred method for investigating complex disease mechanisms in patient-derived organoids, as it can uncover novel, disease-relevant cell states that might not have a distinct morphological signature [91]. The following diagram outlines this decision-making process for researchers.
Validating the composition and health of neural cultures is a critical step in neuroscience research and drug development. The choice of validation method fundamentally influences experimental outcomes, cost, and the very integrity of the precious cellular samples. This guide objectively compares two paradigm approaches: the established, destructive method of immunocytochemistry (ICC) and the emerging, non-destructive method of live-cell morphological profiling. Framed within the broader thesis of improving reproducibility in neural culture research, this analysis provides experimental data and protocols to help researchers select the appropriate workflow for their specific application, particularly when sample preservation is a priority.
Workflow Comparison: A Side-by-Side Analysis
The fundamental difference between these methodologies lies in the fate of the culture post-analysis. Table 1 summarizes the core characteristics of each approach.
Table 1: Core Characteristics of Destructive and Non-Destructive Workflows
| Feature | Destructive (ICC) Workflow | Non-Destructive (Live-Cell Profiling) Workflow |
|---|---|---|
| Sample Integrity | Sample is fixed and stained, ceasing all live functions and dynamics [71]. | Culture remains viable and continues to develop, allowing for longitudinal studies [32] [95]. |
| Key Strength | High molecular specificity for target proteins via antibody-antigen binding [71]. | Reveals dynamic, real-time changes in cell state and function without fixation artifacts [32] [1]. |
| Primary Readout | Snapshots of protein localization and expression at a single time point [71]. | Kinetic data on neurite outgrowth, network formation, and cell movement over time [32] [95]. |
| Throughput | Moderate; limited by multi-step staining and manual imaging. | High; enabled by automation, AI, and continuous imaging inside incubators [32] [95]. |
| Temporal Data | End-point only; requires multiple replicates for time-series data. | Continuous, rich temporal data from a single culture, capturing rare events [32]. |
| Morphology Risk | Potential for artifacts introduced by fixation and permeabilization steps [32]. | Preserves native cell morphology, avoiding fixation-induced alterations [32]. |
The following diagram illustrates the starkly different pathways these workflows take:
Detailed Methodologies and Experimental Protocols
The Destructive Gold Standard: Immunocytochemistry (ICC)
The ICC protocol is a multi-stage process designed to preserve cellular architecture and allow antibody access to intracellular targets.
Sample Preparation and Fixation: Cells are cultured on coverslips or plates, often coated with adhesion-promoting substrates like poly-L-lysine. Fixation, typically with 4% paraformaldehyde (PFA) for 10-20 minutes at room temperature, cross-links proteins to preserve morphology. Alternative fixatives like cold methanol simultaneously fix and permeabilize cells [71].
Permeabilization and Blocking: For PFA-fixed samples, a permeabilization step using detergents like Triton X-100 (0.1-0.2%) is essential to solubilize membranes and allow antibody penetration. This is followed by a blocking step with BSA (2-10%) or serum from the secondary antibody host species to minimize non-specific antibody binding [71].
Antibody Incubation and Imaging: Cells are incubated with a primary antibody specific to the target protein (e.g., Iba1 for microglia, GFAP for astrocytes), followed by a fluorophore-conjugated secondary antibody. After counterstaining for nuclei (e.g., DAPI), the samples are imaged using fluorescence microscopy. The culture is not viable after this process [71] [87].
The Non-Destructive Alternative: Live-Cell Morphological Profiling
This workflow leverages advanced instrumentation and computational analysis to extract rich data from living cultures without harming them.
Live-Cell Imaging Systems: Instruments like the IncuCyte (Sartorius) or similar systems from Agilent, Leica, and others are placed inside standard cell culture incubators. They automatically acquire phase-contrast or fluorescence images at user-defined intervals over days or weeks, maintaining perfect environmental control (37°C, 5% CO₂) [32] [95].
Quantitative Analysis of Neurite Outgrowth: For neuronal networks, a key application is quantifying neurite dynamics. Software algorithms (e.g., NeuroTrack for IncuCyte) automatically segment cell bodies and neurites in phase-contrast images, providing kinetic data on metrics like neurite length per cell and branching points [32].
Cell Painting and AI-Based Classification: A more sophisticated approach uses "Cell Painting," where cells are stained with a panel of non-toxic, fluorescent dyes that target various organelles (nucleus, endoplasmic reticulum, etc.). The resulting morphological "fingerprint" is then analyzed by convolutional neural networks (CNNs) to identify cell types. As demonstrated in mixed neural cultures, this method can discriminate between neurons, neural progenitors, and microglia with over 96% accuracy, all without compromising cell viability [1].
Experimental Data and Performance Comparison
Quantitative Support for Non-Destructive Profiling
Recent studies provide robust experimental data validating the accuracy and utility of non-destructive methods.
- Cerebral Organoid Selection: Ikeda et al. demonstrated that simple morphological classification of cerebral organoids could accurately predict their internal cellular composition, later validated by single-cell RNA sequencing. Organoids with specific morphological features (e.g., "variant 1": rosette-like structures) were confirmed to be primarily composed of cortical glutamatergic neurons, while others were enriched in GABAergic neurons or non-neuronal cells. This allows for the non-destructive selection of desired organoids for experiments, enhancing accuracy and ensuring the safety of cell-based therapies [21].
- Cell-Type Identification in Dense Cultures: An independent study implemented a Cell Painting assay with a CNN classifier to identify cell types in dense, mixed neural cultures derived from iPSCs. The model achieved a remarkable classification accuracy above 96% for distinguishing different neural cell types, showcasing the power of AI-driven morphological profiling as a cheap, simple, and scalable quality control assay [1].
Performance in Functional Assays
Non-destructive imaging truly excels in functional and longitudinal pharmacokinetic studies.
- Real-Time Pharmacodynamic Analysis: Live-cell imaging is indispensable for monitoring the rapid effects of neuroactive compounds. For instance, in hiPSC-derived neuronal networks cultured on micro-electrode arrays (MEAs), drugs like bicuculline (a GABAA receptor antagonist) can be administered while simultaneously monitoring changes in firing rate, network bursting, and functional connectivity in real-time. This provides a dynamic view of drug effects that is impossible to capture with endpoint ICC alone [96].
- Purity Assessment in Microglia Cultures: ICC remains the definitive method for quantifying culture purity post-hoc. For example, in the process of subcloning primary microglia, tertiary cultures were shown by ICC to achieve a purity of 93.1% ± 6.0% (with one attempt reaching ~99%), a critical metric for downstream -omics analysis. However, this confirmation came at the cost of those specific cultures [87].
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 2 details key reagents and instruments used in the workflows discussed in this guide.
Table 2: Key Research Reagent Solutions for Culture Validation
| Item | Function/Application | Example Use-Case |
|---|---|---|
| IncuCyte Systems | Automated live-cell imaging incubator with integrated analysis software. | Kinetic quantification of neurite outgrowth in response to psychoplastogens [32] [95]. |
| Cell Painting Dyes | A panel of fluorescent dyes (e.g., for nucleus, ER, actin) to create a morphological fingerprint. | AI-based identification of cell types (neurons, microglia) in mixed iPSC-derived cultures [1]. |
| Anti-Iba1 Antibody | ICC marker for identifying and quantifying microglia in mixed cultures [87]. | Determining the purity (e.g., ~99%) of a microglia-enriched tertiary culture [87]. |
| Anti-GAD2 / Anti-EMX1 | ICC markers for GABAergic neurons and cortical glutamatergic neurons, respectively [21]. | Validating the cellular composition of cerebral organoids selected by morphology [21]. |
| Poly-L-Lysine | Coating solution for improving adhesion of cells to cultureware. | Preparing coverslips or MEA plates for ICC or live-cell imaging experiments [71] [96]. |
| DPA-TISR Neural Network | Advanced AI for super-resolution live-cell imaging. | Achieving multicolor live-cell SR imaging for >10,000 time points with high fidelity [97]. |
Both destructive and non-destructive workflows are essential tools in the modern neuroscientist's arsenal. ICC remains the gold standard for definitive, high-specificity validation of protein expression and final culture composition. However, the emergence of sophisticated live-cell profiling techniques presents a compelling alternative for kinetic studies, quality control of precious samples, and any experimental pipeline where culture integrity is paramount.
The choice between them is not a matter of which is universally better, but which is optimal for a given research question. For endpoint validation, ICC is unmatched. For understanding dynamic biological processes, preserving unique patient-derived iPSC cultures, or conducting longitudinal drug screens, non-destructive morphological profiling is revolutionizing the field by providing rich, temporal data while keeping the culture alive and functioning.
Multiplexed Protein Localization (ICC) vs. Holistic Phenotypic Capture (Profiling)
This guide provides an objective comparison between two powerful approaches for validating neural cultures: Multiplexed Protein Localization via Immunocytochemistry (ICC) and Holistic Phenotypic Capture via Proteomic Profiling. Understanding the strengths, applications, and data outputs of each method is crucial for selecting the right tool in neuroscience research and drug development.
The functional analysis of neural cultures derived from induced pluripotent stem cells (iPSCs) relies heavily on accurately characterizing cell identity, state, and composition. Two methodological philosophies dominate this space:
- Multiplexed Immunocytochemistry (ICC) is an advanced form of immunostaining that uses antibody-based detection with fluorescent labels to visualize and localize multiple specific protein targets simultaneously within cultured cells. It provides high-resolution, spatial data on protein expression and co-localization within a cellular context. [66] [12]
- Holistic Phenotypic Profiling refers to untargeted, global analysis of cellular components. In the context of this guide, we focus on proteomic profiling, which uses mass spectrometry-based techniques to identify and quantify thousands of proteins in a single experiment, offering a system-wide view of the proteomic landscape without prior selection of targets. [98] [99]
Quantitative Comparison of Technical and Performance Parameters
The choice between these techniques is fundamentally guided by their core performance characteristics, as summarized in the table below.
Table 1: Core performance characteristics of Multiplexed ICC and Proteomic Profiling.
| Parameter | Multiplexed ICC | Holistic Proteomic Profiling |
|---|---|---|
| Primary Readout | Spatial localization and relative abundance of specific proteins | Identification and quantitative abundance of hundreds to thousands of proteins |
| Multiplexing Capacity | 4-12+ targets per sample with sequential staining [100] | >1500 proteins per single experiment [98] [99] |
| Spatial Context | High. Preserves subcellular and single-cell spatial information | None. Typically requires cell lysis, losing all spatial information |
| Sensitivity & Dynamic Range | Medium; semi-quantitative [12] | High; quantitative with broad dynamic range [98] |
| Throughput | Medium to High (compatible with 96-well plates) [66] | Lower; sample processing is more complex and time-consuming |
| Key Advantage | Visual validation of cell identity, state, and protein co-localization | Unbiased discovery of novel biomarkers and pathway alterations |
Experimental Data and Application in Neural Cultures
Insights from Multiplexed ICC
Multiplexed ICC excels at providing spatial validation of neural cultures. A study utilizing PRobe-based Imaging for Sequential Multiplexing (PRISM), an advanced ICC method, on iPSC-derived cortical neurons exemplifies this. The researchers simultaneously imaged 12 protein markers to deconstruct the cellular composition of their cultures with single-cell resolution. [100]
Table 2: Quantitative cell composition analysis of iPSC-derived cortical cultures via multiplexed ICC (PRISM). Data adapted from Tomov et al. 2022. [100]
| Cell Type / State | Defining Markers | Percentage at Day 55 | Percentage at Day 85 |
|---|---|---|---|
| Neurons | MAP2, Tuj1, NeuN | 30% | 58% |
| Excitatory Neurons | VGLUT1 | 100% of identified neurons | Not Specified |
| Radial Glial Cells | Pax6+, Vimentin+ | 36% | Not Specified |
| Immature/Inactive Astrocytes | CD44+, Vimentin+ | 18% | Not Specified |
| Mature/Activated Astrocytes | CD44+, GFAP+ | 7% | Not Specified |
| Neural Progenitor Cells | Pax6+ | 2% | Not Specified |
This data demonstrates the power of multiplexed ICC to track differentiation maturity and heterogeneity over time, providing clear, actionable validation of culture conditions.
Insights from Holistic Proteomic Profiling
In contrast, proteomic profiling provides a global, unbiased view of pathway-level changes. A proteomic study comparing 2D monolayers and 3D neurospheres of hiPSC-derived neural stem cells revealed profound differences. The 3D neurospheres showed enrichment in 542 proteins related to mature neural functions, while 2D monolayers retained a multipotency profile. [98]
Another study used global proteomics to investigate how the loss of the NF1 gene affects human Schwann cells. It identified 148 differentially expressed proteins and specific pathway alterations, including decreased oxidative phosphorylation and increased glycolysis. This holistic approach uncovered a previously underappreciated role for NF1 in mitochondrial metabolism. [99]
Table 3: Key pathways altered in NF1-deficient Schwann cells identified through global proteomics. [99]
| Altered Pathway | Change in NF1-Deficient Cells | Functional Implication |
|---|---|---|
| Oxidative Phosphorylation | Decreased | Reduced mitochondrial energy production |
| Mitochondrial Dysfunction | Increased | Compromised cellular health and viability |
| Glycolysis | Increased | A shift towards less efficient energy metabolism |
| Myelination Signaling | Altered | Implicates NF1 in Schwann cell core function |
Detailed Experimental Protocols
Protocol: PRISM for Multiplexed ICC
The following workflow is adapted from Tomov et al.'s characterization of iPSC-derived cortical and motor neurons. [100]
- Cell Culture and Fixation: Plate dissociated neural cells on coverslips or in imaging chambers. Culture for the desired duration (e.g., 55 vs. 85 days). Fix cells with paraformaldehyde (e.g., 4%) to preserve architecture and permeabilize with a detergent like Triton X-100 to allow antibody entry. [100] [12]
- Antibody Conjugation: Conjugate primary antibodies against neural targets (e.g., MAP2, GFAP, Vimentin) with unique, orthogonal oligonucleotide tags (PRISM probes). [100]
- Staining and Hybridization: Incubate fixed cells with the panel of DNA-conjugated primary antibodies. Then, add fluorescently labeled oligonucleotides that are complementary to the antibody-bound DNA tags. [100]
- Sequential Imaging: Image 3-4 fluorophores at a time using a fluorescence microscope. [100]
- Probe Stripping and Cycling: Gently wash away the fluorescent imaging strands without removing the primary antibodies. This step is non-destructive and does not require harsh antibody stripping. [100]
- Repeat Hybridization and Imaging: Hybridize a new set of fluorescent oligos for the next panel of targets and repeat the imaging process. Cycle through until all 10-12 markers are imaged. [100]
- Image Analysis and Cell Typing: Use automated image analysis software (e.g., CellProfiler) to align images from different cycles and quantify signal. Assign cell identity based on pre-defined marker combinations (see Table 2). [100]
Diagram 1: PRISM multiplexed ICC workflow.
Protocol: TMT-Based Global Proteomic Profiling
The following workflow is adapted from a study comparing 2D and 3D neural cultures and a study on NF1-deficient Schwann cells. [98] [99]
- Sample Preparation: Culture neural cells under different conditions (e.g., 2D vs. 3D, wild-type vs. mutant). Lyse cells to extract total protein. Digest the protein mixture into peptides using an enzyme like trypsin. [98] [99]
- TMT Labeling: Label peptides from each experimental condition with different Tandem Mass Tag (TMT) reagents. These are chemical tags that are isobaric (same mass) but release unique reporter ions during fragmentation, allowing for multiplexed quantification. [98]
- Pooling and Fractionation: Combine the TMT-labeled samples from all conditions into a single tube. To reduce complexity, the peptide pool can be fractionated using high-pH liquid chromatography. [98]
- LC-MS/MS Analysis: Inject the fractionated peptides into a liquid chromatography-mass spectrometry (LC-MS/MS) system. Peptides are separated by chromatography and then ionized and analyzed by the mass spectrometer, which measures peptide mass and sequence. [98]
- Data Analysis and Pathway Mapping: Use bioinformatics software to identify proteins from the mass spectra and quantify their abundance across conditions based on TMT reporter ion intensities. Statistically analyze the data to find differentially expressed proteins and input these into pathway analysis tools (e.g., Ingenuity Pathway Analysis). [98] [99]
Diagram 2: TMT-based proteomic profiling workflow.
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful implementation of these techniques requires specific reagents and tools.
Table 4: Essential materials for multiplexed ICC and proteomic profiling. [100] [98] [99]
| Item | Function/Purpose | Example from Cited Research |
|---|---|---|
| DNA-Conjugated Antibodies | Enable sequential multiplexing in ICC; antibody is linked to a unique DNA barcode. | Custom PRISM antibodies for 12 neural markers (MAP2, GFAP, etc.). [100] |
| Fluorophore-Conjugated Oligos | Complementary imaging strands that bind to antibody barcodes to produce signal. | Locked Nucleic Acid (LNA) or DNA imaging strands. [100] |
| Tandem Mass Tags (TMT) | Isobaric chemical labels for multiplexed quantitative comparison of proteins from different conditions in a single MS run. | 10- or 11-plex TMT kits for comparing 2D vs. 3D neural cultures. [98] |
| Cell Lineage Markers | Antibodies against proteins defining specific neural cell types for ICC validation. | Pax6 (progenitors), Tuj1 (neurons), GFAP (astrocytes). [100] |
| Pathway Analysis Software | Bioinformatics tool to interpret proteomic data and identify altered biological pathways. | Ingenuity Pathway Analysis (IPA) used on NF1 Schwann cell data. [99] |
Multiplexed ICC and holistic proteomic profiling are not mutually exclusive but are powerfully complementary. The choice depends entirely on the research question's stage and scope.
- Choose Multiplexed ICC when your goal is to validate the identity, purity, and spatial organization of neural cultures at a single-cell level. It is ideal for confirming differentiation protocols, characterizing cellular heterogeneity, and confirming the presence of specific, known protein targets in their proper subcellular context.
- Choose Holistic Proteomic Profiling when you need an unbiased, system-wide discovery of differentially expressed proteins and altered pathways. It is the preferred method for identifying novel biomarkers, understanding complex disease mechanisms, and generating new hypotheses without prior assumptions.
For a comprehensive research strategy, proteomic profiling can first identify key altered pathways and protein targets. Subsequently, multiplexed ICC can be deployed to validate these findings and provide spatial context within the complex architecture of neural cultures.
Validating the composition and identity of cells in neural cultures is a critical step in neuroscience research, especially with the increasing use of complex models like those derived from induced pluripotent stem cells (iPSCs). The inherent variability in iPSC differentiation outcomes necessitates robust quality control methods to ensure experimental reproducibility and reliability [1]. Two complementary technologies have emerged as powerful tools for this validation: morphological profiling, an unbiased, high-content screening method, and immunocytochemistry (ICC), a targeted, specificity-driven confirmation technique. Morphological profiling enables the quantitative characterization of cell states based on morphological features, while ICC provides precise molecular identification of specific cell types and proteins.
Each method offers distinct advantages and limitations. The integration of these approaches creates a powerful workflow where morphological profiling can rapidly screen large numbers of samples and conditions, followed by ICC confirmation of specific targets of interest. This integrative approach is particularly valuable for assessing complex mixed neural cultures containing neurons, astrocytes, microglia, and other neural cell types [1] [101]. By leveraging the strengths of both technologies, researchers can achieve comprehensive culture validation with both efficiency and precision.
Technology Comparison: Morphological Profiling vs. Immunocytochemistry
Core Principles and Applications
Morphological Profiling utilizes high-content imaging and computational analysis to quantify hundreds of morphological features from cells, creating a "morphotextural fingerprint" for different cell types and states [1]. Techniques like Cell Painting employ multiplexed fluorescent dyes to label multiple cellular compartments simultaneously, capturing rich morphological data in an unbiased manner [102]. This approach has demonstrated exceptional capability in distinguishing neural cell types, with studies reporting above 96% classification accuracy even in dense, mixed cultures [1]. The method is particularly valuable for detecting subtle phenotypic changes and unexpected effects in neural cultures.
Immunocytochemistry relies on antibody-based detection of specific protein targets, providing high specificity for cell type identification and protein localization. ICC allows researchers to confirm the presence of neuronal markers (e.g., MAP2, TUJ1), synaptic proteins (e.g., synapsin, PSD-95), glial markers (GFAP for astrocytes), and other cell-type-specific proteins [103]. While highly specific, ICC is typically lower in throughput and requires prior knowledge of targets of interest, making it less suitable for discovery-based applications.
Performance Comparison Table
Table 1: Direct comparison of morphological profiling and immunocytochemistry for neural culture validation
| Parameter | Morphological Profiling | Immunocytochemistry |
|---|---|---|
| Classification Accuracy | 96% for neural cell types [1] | >99% for specific protein targets |
| Throughput | High (can process thousands of cells/condition) | Medium to Low (limited by antibody staining and imaging) |
| Multiplexing Capacity | High (6-9 cellular compartments simultaneously with Cell Painting PLUS) [102] | Limited by antibody host species and spectral overlap |
| Cost per Sample | Lower after initial setup | Higher (antibody costs) |
| Technical Expertise | Requires bioinformatics skills | Requires optimization of staining protocols |
| Primary Advantage | Unbiased, discovery-driven | Target-specific, highly precise |
| Best Application | Initial screening, quality control, detecting unexpected phenotypes | Confirmation of specific cell types, protein localization, pathway activation |
Quantitative Performance Data
Table 2: Experimental performance metrics for morphological profiling in neural cultures
| Experimental Context | Classification Performance | Key Findings |
|---|---|---|
| Mixed neural cultures (iPSC-derived) | 96% accuracy distinguishing neurons vs. neural progenitors [1] | Significantly outperformed population-level classification based on time in culture (86%) |
| Cell line models (astrocytoma vs. neuroblastoma) | >96% classification accuracy [1] | Demonstrated unique "morphotextural fingerprints" for each cell type |
| Microglia identification in mixed neuronal cultures | Unequivocal discrimination from neurons [1] | Tiered strategy allowed distinction of activated vs. non-activated states with lower accuracy |
| Convolutional Neural Networks vs. Random Forest | CNN significantly outperformed RF classification [1] | RF classification showed 46% misclassification of specific cell types |
Methodological Protocols
Morphological Profiling Workflow for Neural Cultures
The Cell Painting protocol provides a standardized approach for morphological profiling [102] [104]. For neural cultures, specific adaptations enhance performance:
Sample Preparation:
- Plate neural cultures in appropriate vessels compatible with high-content imaging systems
- Maintain appropriate cell densities to ensure proper segmentation while preserving physiological relevance
- Include appropriate controls (known cell types, treatment conditions) for assay validation
Staining Protocol (Cell Painting):
- Fix cells with 4% paraformaldehyde for 15 minutes at room temperature
- Permeabilize with 0.1% Triton X-100 for 10-15 minutes
- Stain with the following dye cocktail:
- Nuclear DNA: Hoechst 33342 (1-5 µg/mL)
- Cytoplasmic RNA: SYTO RNASelect (500 nM)
- Endoplasmic Reticulum: Concanavalin A, Alexa Fluor 488 conjugate (100 µg/mL)
- Mitochondria: MitoTracker Deep Red (100 nM)
- Actin Cytoskeleton: Phalloidin (various fluorophores, 200-400 nM)
- Golgi Apparatus: Anti-Golgin-97 antibody [102]
- Incubate for appropriate times based on dye penetration requirements
- Wash thoroughly to remove non-specific staining
Image Acquisition and Analysis:
- Acquire images using high-content microscopes with appropriate magnification (20x-60x)
- Ensure sufficient cells per condition (minimum 500-1000 cells for robust statistics)
- Use automated image analysis pipelines (CellProfiler, ImageJ) for segmentation and feature extraction [104]
- Extract features describing morphology, texture, intensity, and spatial relationships
- Apply machine learning classifiers (CNN, Random Forest) for cell type identification [1]
Immunocytochemistry Confirmation Protocol
Sample Preparation:
- Culture neural cells on coverslips or in imaging-compatible plates
- Fix with 4% PFA for 15 minutes at room temperature
- Permeabilize with 0.3% Triton X-100 for 15 minutes
- Block with 5% normal serum from secondary antibody host species for 1 hour
Antibody Staining:
- Prepare primary antibodies in blocking solution at optimal concentrations:
- Incubate overnight at 4°C or 2 hours at room temperature
- Wash 3x with PBS (5 minutes each)
- Incubate with species-appropriate secondary antibodies (1:500-1:1000) for 1 hour at room temperature, protected from light
- Counterstain with DAPI (1 µg/mL) for 5 minutes
- Mount with anti-fade mounting medium
Image Acquisition and Analysis:
- Acquire images using confocal or epifluorescence microscopy
- Ensure appropriate controls (no primary antibody, isotype controls)
- Use consistent exposure settings across comparisons
- Quantify marker expression using image analysis software
Integrated Workflow Visualization
Integrated Validation Workflow: This diagram illustrates the sequential integration of morphological profiling for initial screening followed by immunocytochemistry for targeted confirmation in neural culture validation.
Neural Culture Composition and Marker Expression
Neural Culture Components: This diagram shows the major cell types present in mixed neural cultures and their corresponding identification markers used in morphological profiling and immunocytochemistry.
Research Reagent Solutions
Table 3: Essential research reagents for morphological profiling and ICC in neural culture validation
| Reagent Category | Specific Examples | Function & Application |
|---|---|---|
| Cell Painting Dyes | Hoechst 33342, SYTO RNASelect, Concanavalin A, MitoTracker, Phalloidin [102] | Multiplexed staining of cellular compartments for morphological profiling |
| Neuronal Markers | βIII-tubulin (TUJ1), MAP2, NeuN, Neurofilament [103] [101] | ICC identification of neuronal cells and processes |
| Glial Markers | GFAP (astrocytes), IBA1 (microglia), Olig2 (oligodendrocytes) [1] [105] | ICC identification of specific glial cell types |
| Synaptic Markers | PSD-95, Synapsin, Synaptophysin, VGLUT1 [103] | ICC validation of synaptic structures and function |
| Proliferation Markers | Ki67, pH3, EdU [101] | ICC identification of dividing neural progenitor cells |
| Fixation Reagents | 4% Paraformaldehyde, Methanol [102] [104] | Cell preservation for both morphological and ICC analysis |
| Permeabilization Agents | Triton X-100, Tween-20, Saponin [104] | Enable antibody and dye access to intracellular targets |
| Blocking Reagents | Normal serum, BSA, Fish skin gelatin [101] | Reduce non-specific antibody binding in ICC |
| Mounting Media | ProLong Diamond, Vectashield with DAPI [101] | Preserve fluorescence and provide nuclear counterstain |
Advanced Applications and Future Directions
Three-Dimensional and Complex Neural Systems
As neural culture systems advance toward more physiologically relevant three-dimensional models, including brain organoids and neurospheroids, validation approaches must adapt accordingly [43] [105]. Morphological profiling faces challenges in 3D environments due to light scattering and segmentation complexities, but advances in light-sheet microscopy and computational methods are addressing these limitations. Researchers have successfully applied live imaging to track brain organoid development over weeks, monitoring tissue morphology, cell behaviors, and subcellular features [43]. For 3D systems, ICC validation becomes more challenging due to antibody penetration issues, often requiring sectioning or specialized clearing techniques.
Computational Advances in Morphological Analysis
Deep learning approaches are revolutionizing morphological profiling by enabling accurate segmentation and quantification even with imperfect training data [106]. These advances are particularly valuable for neural cultures with complex morphologies such as extensive neurite outgrowth and branching. Convolutional neural networks (CNNs) have demonstrated superior performance compared to traditional random forest classifiers for neural cell type identification, achieving above 96% accuracy in distinguishing different neural cell types [1]. The development of approaches that work with imperfect annotations significantly reduces the preparation time for training data, making deep learning more accessible for neural culture validation [106].
High-Content Screening in Drug Discovery
The integration of morphological profiling with ICC confirmation provides a powerful platform for drug screening using neural cultures. For example, researchers have utilized this approach to assess the effects of anti-seizure medications on iPSC-derived neuronal cultures, demonstrating compound-specific responses in both morphological and functional assays [103]. This integrated validation approach ensures that observed drug effects can be properly attributed to specific cell types and states within complex mixed neural cultures, enhancing the reliability of screening outcomes.
The integration of morphological profiling for initial screening followed by immunocytochemistry for target confirmation represents a powerful paradigm for neural culture validation. This combined approach leverages the strengths of both methods: the unbiased, high-throughput capability of morphological profiling to identify cell types and detect unexpected phenotypes, and the high specificity of ICC to confirm critical targets and provide molecular validation. Experimental data demonstrates that morphological profiling can achieve above 96% accuracy in classifying neural cell types, while ICC provides the essential ground truth for specific protein markers.
As neural culture models continue to increase in complexity, from 2D monolayers to 3D organoids and brain-on-chip systems, robust validation strategies become increasingly critical. The integrated approach outlined here provides researchers with a comprehensive framework for ensuring culture quality, composition, and reproducibility. By implementing this combined workflow, neuroscience researchers and drug development professionals can enhance the reliability of their models and accelerate the discovery process for neurological disorders and treatments.
In the field of neural culture research, the ability to accurately identify and characterize cell types is a fundamental prerequisite for reliable disease modeling and drug discovery. The core challenge lies in the inherent variability and density of mixed neural cultures derived from induced pluripotent stem cells (iPSCs). Two dominant techniques have emerged for addressing this validation challenge: traditional immunocytochemistry (ICC), which relies on specific antibody-based protein detection, and emerging AI-powered morphological profiling, which uses high-content imaging and machine learning to identify cell types based on visual features alone. This guide provides an objective comparison of these methodologies, evaluating their performance, requirements, and applicability for researchers validating neural culture composition.
The adoption of iPSC technology has revolutionized cell biology by enabling the generation of human neurons, astrocytes, microglia, and other brain-resident cells. However, genetic drift, clonal heterogeneity, and protocol variations cause significant variability in differentiation outcomes, directly impacting experimental reproducibility [107]. This cellular heterogeneity challenges correct data allocation and interpretation, creating a pressing need for robust validation methods that can ensure culture purity and composition [101].
Table: Core Challenge Comparison in Neural Culture Validation
| Aspect | Immunocytochemistry (ICC) | AI-Powered Morphological Profiling |
|---|---|---|
| Primary Basis | Antibody-antigen recognition of specific protein markers | Pattern recognition of cellular and subcellular structures |
| Throughput Potential | Lower (manual processing, limited multiplexing) | Higher (automated, high-content imaging) |
| Quantitative Capability | Semi-quantitative (intensity-based) | Highly quantitative (multi-parametric analysis) |
| Technical Variability | High (antibody lot variability, staining protocols) | Moderate (imaging conditions, model training) |
| Cost Structure | High reagent costs, labor-intensive | High initial infrastructure, lower marginal cost |
Technical Comparison: Performance Metrics and Experimental Data
Accuracy and Classification Performance
Direct comparative studies reveal significant differences in classification accuracy between the two approaches. In controlled studies using dense mixed neural cultures, AI-powered morphological profiling demonstrated exceptional capability in discriminating neural cell types. One study implementing cell painting with convolutional neural networks (CNNs) achieved a classification accuracy above 96% when benchmarking using pure and mixed cultures of neuroblastoma and astrocytoma cell lines [107].
The same study found that morphological single-cell profiling significantly outperformed time-in-culture as a classification criterion (96% vs. 86%), demonstrating that AI-based analysis surpasses even temporal developmental cues in identifying cell states [107]. Furthermore, in mixed iPSC-derived neuronal cultures, microglia could be "unequivocally discriminated from neurons, regardless of their reactivity state" using this approach [107].
While immunocytochemistry remains the gold standard for specific marker identification, its accuracy is highly dependent on antibody specificity and staining quality. Studies have noted that ICC-based approaches face challenges with cellular heterogeneity, where the "development of non-iSN cells in these cultures remains an issue" despite protocol optimization [101].
Processing Time and Workflow Efficiency
AI-powered methods demonstrate substantial advantages in processing speed, particularly for large-scale studies. The Digital PATH Project, which compared 10 digital pathology tools, highlighted that the evaluation of 1,100 breast cancer samples was completed "in a matter of days and weeks" using AI approaches, representing a highly efficient validation process [108].
In practical clinical applications, tools like Nuclei.io have demonstrated remarkable time savings, with tasks that normally take pathologists "five minutes to 10 minutes" reduced to "a few seconds" with AI assistance [109]. This efficiency gain translates directly to research settings, where AI can rapidly screen large culture sets.
Immunocytochemistry requires extensive processing time including fixation, permeabilization, antibody incubation, and washing steps, typically requiring 24-48 hours for complete processing before analysis can even begin.
Table: Time Investment Comparison for Neural Culture Validation
| Process Step | Immunocytochemistry | AI Morphological Profiling |
|---|---|---|
| Sample Preparation | 2-4 hours (fixation, permeabilization) | 30 minutes (staining with simple dyes) |
| Primary Processing | 12-16 hours (antibody incubation) | 1-2 hours (imaging acquisition) |
| Analysis Phase | 1-2 hours (manual counting/imaging) | Minutes (automated algorithm processing) |
| Total Hands-on Time | 3-6 hours | 1-2 hours |
| Total Elapsed Time | 24-48 hours | 3-6 hours |
Multiplexing Capability and Data Richness
Morphological profiling excels in multiparametric data extraction from single samples. The cell painting approach utilizes multiple fluorescent channels to capture diverse cellular components, generating hundreds of quantitative features describing shape, intensity, texture, and spatial relationships [107]. One study extracted features "for each channel in three regions of interest, namely the nucleus, cytoplasm and whole cell," describing "shape, intensity and texture features of each ROI" [107].
Immunocytochemistry faces practical limitations in multiplexing due to antibody host species limitations and spectral overlap. While sequential staining approaches exist, they increase processing time and potential sample damage. AI methods can continuously expand their analytical capabilities by training on new data, creating a self-improving cycle where "pathologists can share their models with colleagues—almost like a social network" to enhance diagnostic power [109].
Experimental Protocols and Methodologies
AI-Powered Morphological Profelling Protocol
The following protocol for AI-powered cell type validation in neural cultures is adapted from published research demonstrating 96% classification accuracy [107]:
Sample Preparation and Staining:
- Culture cells on appropriate imaging-compatible surfaces (e.g., glass coverslips or multi-well plates)
- Fix cells with 4% paraformaldehyde for 15 minutes at room temperature
- Permeabilize with 0.1% Triton X-100 for 10 minutes
- Implement Cell Painting staining cocktail:
- Nuclear stain: Hoechst 33342 (1-5 µg/mL) for 30 minutes
- Cytoplasmic stain: Phalloidin (conjugated to Alexa Fluor 488, 1:200) for 30 minutes
- Mitochondrial stain: MitoTracker Deep Red (100-500 nM) for 30 minutes
- Nucleolar stain: SYTO 14 green fluorescent nucleic acid stain (1-5 µM) for 30 minutes
- Endoplasmic reticulum stain: Concanavalin A (conjugated to Alexa Fluor 647, 100 µg/mL) for 30 minutes
- Wash with PBS and maintain in appropriate mounting medium
Image Acquisition and Processing:
- Acquire images using high-content confocal microscope with 20x or 40x objective
- Capture z-stacks (5-7 slices at 0.5-1 µm interval) for each channel
- Use automated image analysis pipeline:
- Cell segmentation using nuclear marker as primary mask
- Cytoplasmic expansion using watershed algorithm
- Feature extraction for each cell (1,500+ morphological features)
- Data normalization and quality control
AI Model Training and Classification:
- Partition data into training (70%), validation (15%), and test (15%) sets
- Train convolutional neural network (ResNet architecture recommended) using labeled data
- Validate model performance using independent test set
- Apply model to new images for cell type prediction
- Implement visualization techniques (Grad-CAM) for model interpretability
AI-Powered Morphological Profiling Workflow
ICC Culture Purity Optimization Protocol
This protocol for improving culture purity using ICC-based methods incorporates cytostatic treatment to reduce non-neuronal cells, adapted from published optimization studies [101]:
Differentiation and Treatment:
- Differentiate iSNs from iPSC line using established small-molecule inhibition protocol
- Seed cells on growth factor-reduced Matrigel-coated coverslips on day +10 of differentiation
- Apply floxuridine (FdU) treatment:
- Prepare 10 μM FdU in neuronal maturation medium
- Treat cultures for 24 hours
- Perform half-medium change to ensure stable FdU concentration for precise timing
- Include control groups without FdU treatment for comparison
Immunostaining and Analysis:
- Fix cells with 4% PFA for 15 minutes at room temperature
- Permeabilize with 0.3% Triton X-100 for 10 minutes
- Block with 5% normal goat serum for 1 hour
- Apply primary antibodies for neuronal markers:
- βIII-tubulin (1:1000) for pan-neuronal identification
- MAP2 (1:500) for mature neurons
- NeuN (1:500) for post-mitotic neurons
- GFAP (1:1000) for astrocytes
- Iba1 (1:800) for microglia
- Incubate overnight at 4°C
- Apply appropriate secondary antibodies (1:1000) for 2 hours at room temperature
- Counterstain with DAPI (1 µg/mL) for 10 minutes
- Image using epifluorescence or confocal microscopy
- Quantify cell counts using automated or manual methods
Validation and Quality Control:
- Assess cell viability using luminescent assay
- Calculate iSN-to-total-cell-count ratio
- Evaluate neuronal functionality using multi-electrode array measurements
- Confirm sensory neuron-specific marker expression (e.g., peripherin, TRPV1)
ICC Culture Purity Optimization Workflow
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table: Key Research Reagents for Neural Culture Validation
| Reagent/Solution | Function | Example Application |
|---|---|---|
| Floxuridine (FdU) | Cytostatic compound targeting proliferating non-neuronal cells | Culture purity optimization at 10μM for 24h treatment [101] |
| Cell Painting Cocktail | Multi-channel fluorescent staining for morphological profiling | Simultaneous labeling of multiple organelles for AI-based classification [107] |
| Matrigel-Coated Surfaces | Basement membrane matrix for cell attachment and differentiation | Providing optimal substrate for iPSC differentiation and neuronal maturation [101] |
| Neural Crest Stem Cell MicroBeads | Magnetic-activated cell sorting for neuronal population enrichment | Isolation of specific neural cell types from mixed cultures [101] |
| N2B27 Medium | Defined medium for neuronal maturation and maintenance | Supporting long-term culture of differentiated neurons [101] |
| Y27632 (ROCK inhibitor) | Enhances cell survival after passaging | Improving viability during subculture of neural populations [101] |
| LDN-193189/SB-431542 | Small molecule inhibitors for neural induction | Directing iPSC differentiation toward neural lineages [101] |
| BDNF/GDNF/NGF | Neurotrophic factors supporting neuronal health | Enhancing maturation and functional activity of iSNs [101] |
The comparison between immunocytochemistry and AI-powered morphological profiling reveals complementary strengths that researchers can strategically leverage based on specific validation requirements. ICC provides high specificity for known markers and remains essential for confirming expression of specific proteins, while morphological profiling offers unparalleled efficiency for high-content screening and discovery of novel morphological patterns.
For researchers future-proofing their validation approaches, AI-powered methods demonstrate clear advantages in scalability, quantitative power, and adaptability to new questions. The field is rapidly evolving toward integrated approaches where "AI can now quantify complex cellular interactions within the tumor microenvironment, providing more nuanced predictions about treatment response" [110]. This capability translates directly to neural culture validation, where understanding complex cell-cell interactions will be crucial for advanced disease modeling.
The most forward-looking approaches will likely combine both techniques—using ICC for definitive marker confirmation while implementing AI-powered morphological profiling for routine quality control and novel discovery. As the technology continues to mature, "digital pathology and AI will likely become the new standard of care, driving higher efficiency, improved patient outcomes, and deeper scientific understanding" [111] in both clinical and research contexts. Researchers who adopt and master these integrated approaches now will be best positioned to leverage continuing advancements in digital pathology and artificial intelligence.
Conclusion
The comparative analysis reveals that morphological profiling and immunocytochemistry are not mutually exclusive but are powerfully complementary. Morphological profiling, supercharged by AI and live-cell imaging, offers an unparalleled, non-destructive tool for high-throughput screening, dynamic monitoring, and reducing batch variability in neural cultures. Immunocytochemistry remains the gold standard for definitive, high-specificity molecular validation. The future of neural culture validation lies in integrated workflows that leverage the speed and scalability of morphological profiling for initial quality control and experimental triage, followed by targeted ICC for deep mechanistic insight. This synergistic approach, supported by standardized quality control frameworks and advancing digital pathology, will be crucial for accelerating the development of more reproducible disease models, effective neurotherapeutics, and safe cell-based transplantation therapies.
References
- 1. Unbiased identification of cell identity in dense mixed ... [https://elifesciences.org/articles/95273]
- 2. Morphological, immunocytochemical, and functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4520575/]
- 3. The NERVE-ML (neural engineering reproducibility and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11948487/]
- 4. A brain-wide map of neural activity during complex behaviour [https://www.nature.com/articles/s41586-025-09235-0]
- 5. MIT researchers invent new human brain model to enable ... [https://news.mit.edu/2025/researchers-invent-human-brain-model-to-enable-disease-research-drug-discovery-1105]
- 6. Co-cultured sensory neuron classification using ... [https://pubmed.ncbi.nlm.nih.gov/41038255/]
- 7. What is Immunocytochemistry (ICC)? [https://www.sinobiological.com/category/what-is-icc]
- 8. Immunocytochemistry [https://grokipedia.com/page/Immunocytochemistry]
- 9. Brief guide to immunostaining - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11699723/]
- 10. Morphological and immunocytochemical characteristics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2707099/]
- 11. Comparison of 2D and 3D neural induction methods for the ... [https://www.sciencedirect.com/science/article/pii/S1873506117302131]
- 12. Immunocytochemistry/Immunofluorescence Guide - ICC/IF ... [https://www.antibodies.com/applications/immunofluorescence]
- 13. Immunocytochemistry and fluorescence imaging efficiently ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5540436/]
- 14. Abcam: Leading the way in antibody performance [https://www.abcam.com/en-us/stories/articles/ycharos-results-of-abcam-and-competitor-antibodies]
- 15. Combining phenomics with transcriptomics reveals cell-type ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12241582/]
- 16. In vitro morphological profiling of T cells predicts clinical ... [https://www.nature.com/articles/s41467-025-60224-3]
- 17. Structural and molecular differentiation of cultured human ... [https://www.nature.com/articles/s41598-025-15561-0]
- 18. Systematic Quantification of Synapses in Primary Neuronal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7516133/]
- 19. Immunocytochemical characterization of neuron-ricprimary ... [https://www.sciencedirect.com/science/article/pii/0006899386910061]
- 20. Long-range directional growth of neurites induced by ... [https://www.sciencedirect.com/science/article/pii/S1742706124007773]
- 21. Validation of non-destructive morphology-based selection ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11589179/]
- 22. DELTA: a method for brain-wide measurement of synaptic ... [https://www.nature.com/articles/s41593-025-01923-4]
- 23. Stem Cells and Organoids: A Paradigm Shift in Preclinical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12299584/]
- 24. iPSC-derived and Patient-Derived Organoids: Applications ... [https://www.sciencedirect.com/science/article/pii/S2666027X24000501]
- 25. Morphological profiling for drug discovery in the era of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11182685/]
- 26. AI-based virtual immunocytochemistry for rapid and robust ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12273370/]
- 27. Self-supervision advances morphological profiling by ... [https://www.nature.com/articles/s41598-025-88825-4]
- 28. Induced pluripotent stem cells (iPSCs) [https://www.nature.com/articles/s41392-024-01809-0]
- 29. Examining the Market for Organoid Culture Systems in 2025 [https://eureka.patsnap.com/report-examining-the-market-for-organoid-culture-systems-in-2025]
- 30. A Refined Culture System for Human Induced Pluripotent ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5768885/]
- 31. Live-Cell Immunocytochemistry [https://www.sartorius.com/en/applications/life-science-research/live-cell-assays/live-cell-immunocytochemistry]
- 32. Real-Time Analysis of Neuronal Cell Cultures for CNS ... [https://www.mdpi.com/2076-3425/14/8/770]
- 33. Live Cell Imaging, Live Cell Applications, Time-Lapse ... [https://www.moleculardevices.com/applications/live-cell-imaging]
- 34. Live Cell in Vitro and in Vivo Imaging Applications [https://pmc.ncbi.nlm.nih.gov/articles/PMC3864231/]
- 35. Live-Cell Imaging and Analysis System: Incucyte ® S3 [https://www.sartorius.com/en/products/live-cell-imaging-analysis/live-cell-analysis-instruments/s3-live-cell-analysis-instrument]
- 36. Live Cell Imaging & Analysis [https://www.sartorius.com/en/products/live-cell-imaging-analysis]
- 37. A Comparison of Real-Time and Endpoint Cell Viability ... [https://www.sciencedirect.com/science/article/pii/S247255522207294X]
- 38. Adaptations to the neuronal culture for researchers at ... [https://www.sciencedirect.com/science/article/abs/pii/S0165027025000780]
- 39. CelloType: a unified model for segmentation and ... [https://www.nature.com/articles/s41592-024-02513-1]
- 40. Optimized CNN framework for malaria detection using Otsu ... [https://www.nature.com/articles/s41598-025-23961-5]
- 41. CNN based method for classifying cervical cancer cells in ... [https://www.nature.com/articles/s41598-025-10009-x]
- 42. Advancing blood cell detection and classification [https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-025-03027-2]
- 43. Morphodynamics of human early brain organoid ... [https://www.nature.com/articles/s41586-025-09151-3]
- 44. Segmentation Matters: Recognizing the Cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12407710/]
- 45. ICC vs IHC vs IF ? Do You Know The Difference? [https://www.bio-techne.com/applications/imaging/immunohistochemistry/icc-ihc-if]
- 46. Morphological profiling in human neural progenitor cells ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10994270/]
- 47. Evolution and impact of high content imaging [https://www.sciencedirect.com/science/article/pii/S2472555223000667]
- 48. High-content analysis of neuronal morphology in human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9386098/]
- 49. Unbiased identification of cell identity in dense mixed ... [https://elifesciences.org/reviewed-preprints/95273v1]
- 50. High-Content 3D Assay for Morphological Characterization ... [https://www.moleculardevices.com/en/assets/app-note/dd/img/high-content-assay-for-morphological-characterization-of-neuronal-development-in-3d-matrix-using-human-ipsc-derived-neuronal-cultures]
- 51. Applications of High Content Screening in Life Science ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2841426/]
- 52. Automated quantification of synapses by fluorescence ... [https://www.sciencedirect.com/science/article/abs/pii/S0165027011006868]
- 53. Quantifying Synapses: an Immunocytochemistry-based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3159596/]
- 54. Synaptic markers for pre- and postsynaptic regions [https://www.alomone.com/synaptic-markers-for-pre-and-postsynaptic-regions?srsltid=AfmBOopHZ92LKX-RpaIlmZMKbMEnXRssbIti_rJ8gO0IGMPI5a-Ta-sO]
- 55. Modeling neurodegenerative diseases with brain organoids [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1663286/full]
- 56. Stages of neuronal morphological development in vitro— ... [https://www.sciencedirect.com/science/article/abs/pii/S0165027011002536]
- 57. Morphological and Molecular Profiling of Amyloid-β Species in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11880078/]
- 58. Morphology of Alzheimer's disease and related disorders [https://link.springer.com/chapter/10.1007/978-3-7091-3396-5_6]
- 59. Alzheimer and Parkinson are two faces of the same disease [https://www.sciencedirect.com/science/article/pii/S2667242122000690]
- 60. Comparative analysis of neuroinflammatory pathways in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12069400/]
- 61. Brain organoids-on-chip for neural diseases modeling [https://www.sciencedirect.com/science/article/pii/S2095177925001406]
- 62. Towards a quality control framework for cerebral cortical ... [https://www.nature.com/articles/s41598-025-14425-x]
- 63. Towards a quality control framework for cerebral cortical ... [https://netri.com/towards-a-quality-control-framework-for-cerebral-cortical-organoids-netris-new-publication-in-nature-scientific-reports/]
- 64. Towards a quality control framework for cerebral cortical organoids - PubMed [https://pubmed.ncbi.nlm.nih.gov/40790335/]
- 65. New Quality Control Framework for Cerebral Organoids Can Help Scale Their Application [https://www.pcrm.org/news/innovative-science/new-quality-control-framework-cerebral-organoids-can-help-scale-their]
- 66. Comparing IHC, ICC, and IF: Which One Fits Your Research? [https://www.creative-bioarray.com/comparing-ihc-icc-and-if-which-one-fits-your-research.htm]
- 67. On the Necessity of Validating Antibodies ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6497795/]
- 68. Antibody Validation: Ensuring Quality and Reliability [https://www.sysy.com/antibody-validation]
- 69. The Importance of IHC/ICC Controls [https://www.rndsystems.com/resources/protocols/importance-ihcicc-controls]
- 70. Immunocytochemistry: Human Neural Stem Cells - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2565851/]
- 71. Immunocytochemistry protocol [https://www.abcam.com/en-us/technical-resources/protocols/icc-protocol]
- 72. Human primary mixed brain cultures - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC4181361/]
- 73. Optical segmentation-based compressed readout of ... [https://www.nature.com/articles/s41467-025-62663-4]
- 74. Evaluating feature extraction in ovarian cancer cell line co ... [https://www.nature.com/articles/s42003-025-07766-w]
- 75. A modular framework for multi-scale tissue imaging and ... [https://www.nature.com/articles/s41467-024-48146-y]
- 76. Morphological profiling data resource enables prediction of ... [https://www.sciencedirect.com/science/article/pii/S2589004225007060]
- 77. Strategies to overcome the limitations of current organoid ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12033597/]
- 78. Breakthroughs and challenges of organoid models for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12059183/]
- 79. Label-free in-line characterization of immune cell culture ... [https://www.nature.com/articles/s41536-025-00442-x]
- 80. Linking autism risk genes to morphological and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12319652/]
- 81. High-Dimensional Gene Expression and Morphology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10012424/]
- 82. Comparison of Immunocytochemistry and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5015499/]
- 83. Morphological subprofile analysis for bioactivity annotation ... [https://www.sciencedirect.com/science/article/pii/S2451945623001599]
- 84. Immunocytochemistry - an overview [https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/immunocytochemistry]
- 85. Establishing predictive machine learning models for drug ... [https://www.nature.com/articles/s41698-025-00937-2]
- 86. The principle and method of Immunocytochemistry (IC) [https://www.mblbio.com/bio/g/support/method/immunocytochemistry.html]
- 87. Quantitative morphometric and cell-type-specific ... [https://www.sciencedirect.com/science/article/pii/S2667242121000087]
- 88. Immunohistochemistry for Pathologists: Protocols, Pitfalls, ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5122731/]
- 89. Neuronal Cell Culture Media Growth Opportunities and ... [https://www.datainsightsmarket.com/reports/neuronal-cell-culture-media-1767363]
- 90. Validation of non-destructive morphology-based selection ... [https://www.sciencedirect.com/science/article/pii/S2213671124002674]
- 91. An integrated transcriptomic cell atlas of human neural ... [https://www.nature.com/articles/s41586-024-08172-8]
- 92. Optimized Protocol for Culture of Mouse Fetal Hindbrain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12153581/]
- 93. Generation and validation of a iPSC line from a healthy ... [https://www.sciencedirect.com/science/article/pii/S1873506125001941]
- 94. Navigating Brain Organoid Maturation: From Benchmarking ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12383812/]
- 95. Real-Time Analysis of Neuronal Cell Cultures for CNS Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11352746/]
- 96. In vitro electrophysiological drug testing on neuronal networks ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-024-04018-2]
- 97. A neural network for long-term super-resolution imaging of ... [https://www.nature.com/articles/s41587-025-02553-8]
- 98. A fast TMT-based proteomic workflow reveals neural ... [https://pubmed.ncbi.nlm.nih.gov/41253251/]
- 99. Global proteomics and affinity mass spectrometry analysis ... [https://www.nature.com/articles/s41598-024-84493-y]
- 100. Resolving cell state in iPSC-derived human neural ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8225800/]
- 101. Human sensory-like neuron cultivation—An optimized ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11484056/]
- 102. Cell Painting PLUS: expanding the multiplexing capacity of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12022024/]
- 103. Drug treatment alters performance in a neural ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12174357/]
- 104. Data-analysis strategies for image-based cell profiling [https://www.nature.com/articles/nmeth.4397]
- 105. Exploring neuronal circuitry in neurodegenerative diseases [https://pubs.rsc.org/en/content/articlehtml/2025/lc/d5lc00125k]
- 106. Quantifying morphologies of developing neuronal cells ... [https://www.sciencedirect.com/science/article/pii/S2667242123022959]
- 107. Unbiased identification of cell identity in dense mixed ... [https://elifesciences.org/reviewed-preprints/95273]
- 108. Diagnostics World - Comparison of AI Digital Pathology ... [https://friendsofcancerresearch.org/news/diagnostics-world-comparison-of-ai-digital-pathology-tools-finds-means-to-measure-performance/]
- 109. AI tool gives pathologists speed, accuracy and a new way to ... [https://med.stanford.edu/news/all-news/2025/09/ai-tool-pathology.html]
- 110. Digital Pathology and AI Highlights from ASCO 2025 [https://proscia.com/digital-pathology-and-ai-highlights-from-asco-2025/]
- 111. Digital Pathology and Artificial Intelligence 2025 [https://mdforlives.com/blog/digital-pathology-and-artificial-intelligence/]